Bacillus subtilis: Difference between revisions
imported>Annette Kosiorowski |
mNo edit summary |
||
| (28 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{subpages}} | {{subpages}} | ||
{{Taxobox | {{Taxobox | ||
| color = pink | | color = pink | ||
| Line 16: | Line 14: | ||
| binomial_authority = | | binomial_authority = | ||
}} | }} | ||
===Description and significance=== | ===Description and significance=== | ||
''Bacillus subtilis'' is a [[ | ''Bacillus subtilis'' is a [[Gram-positive]], rod-shaped, [[endospore]]-forming [[bacteria]]. It is regarded as an [[aerobe]]; however, it is also capable of growing and sporulating under [[anaerobic]] conditions when necessary. ''B. subtilis'' resides predominantly in soil, including low-nutrient soil. Due to its association with soil particles, it is also inevitably transferred to plants, foods, animals and even marine and freshwater habitats. | ||
''B. subtilis'' was one of the first bacteria studied by scientists. It was originally named ''Vibrio subtilis'' in 1835 by [[Christian Gottfried Ehrenberg]] and later renamed ''Bacillus subtilis'' by [[Ferdinand Cohn]] in 1872. | ''B. subtilis'' was one of the first bacteria studied by scientists. It was originally named ''Vibrio subtilis'' in 1835 by [[Christian Gottfried Ehrenberg]] and later renamed ''Bacillus subtilis'' by [[Ferdinand Cohn]] in 1872. | ||
| Line 25: | Line 22: | ||
==Genome structure== | ==Genome structure== | ||
The genome sequence of ''Bacillus subtilis'' was completed and published in November 1997.<ref>[http://bacillus.genome.jp | B. subtilis Genome]</ref><ref>[http://www.nature.com/nature/journal/v390/n6657/full/390249a0.html "The complete genome sequence of the Gram-positive bacterium Bacillus subtilis."]</ref> It was the first Gram-positive bacteria to have its genome sequenced. ''B. subtilis'' contains a single, circular [[chromosome]]. It consists of 4,214,810 base pairs. The average G+C content is 43.5%, although ten regions have a G+C content significantly lower than average. There is a significant G-T skew at the third [[codon]] positions in common with other bacteria. Its leading strand has a 9% excess of G and 4% excess of T, as compared to the lagging strand. Approximately 87% of the genome consists of protein-coding regions. More than 74% of the [[open reading frames]] and 94% of ribosomal genes are transcribed co-directionally with replication. Only 53% of the genes are represented once. The remaining belong to [[multigene families]], which range in size from 2 to 77 gene copies. Approximately 220 transcriptional regulators have been identified in the genome. At the present time, the functions of only approximately 58% of the genes are known, while the functions of 42% are still unknown and being studied. Twelve percent of these genes with unknown functions have homologs in other organisms.<ref>Devine, Kevin M. (2000). "Bacillus subtilis: Genetics."</ref> | |||
[[Replication]] of the ''B. subtilis'' genome proceeds similarly to most bacteria, with the splitting of the chromosome into two replication forks. ''B. subtilis'' has an origin of replication (oriC) and a terminus of replication (terC) located almost perfectly opposite each other on the genome. However, some studies have shown that the clockwise and counterclockwise replication forks actually differ in length by about 170 base pairs. Thus, once the clockwise replication fork reaches the terminus, its synthesis is terminated, while synthesis of the counterclockwise fork continues until it finally reaches the terminus. .<ref>Lonenshein, A.L., Hoch, J.A., & Losick, R. (1993). ''Bacillus subtilis and other Gram-positive bacteria.''</ref> | |||
[[Replication]] of the ''B. subtilis'' genome proceeds similarly to most bacteria, with the splitting of the chromosome into two replication forks. ''B. subtilis'' has an origin of replication (oriC) and a terminus of replication (terC) located almost perfectly opposite each other on the genome. However, some studies have shown that the clockwise and counterclockwise replication forks actually differ in length by about 170 base pairs. Thus, once the clockwise replication fork reaches the terminus, its synthesis is terminated, while synthesis of the counterclockwise fork continues until it finally reaches the terminus. | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
''Bacillus subtilis'' is a [[prokaryotic]] cell, lacking membrane-bound organelles. It is enclosed by a cell wall, consisting of large amounts of [[peptidoglycan]] (Gram-positive). It is rod, or [[bacillus]], shaped and usually found in colonies with other ''B. subtilis''. ''B. subtilis'' is flagellated and thus motile. | |||
''Bacillus subtilis'' is a [[prokaryotic]] cell, lacking membrane-bound organelles. It is enclosed by a cell wall, consisting of large amounts of [[peptidoglycan]] ( | |||
[[Cellular respiration]] in ''B. subtilis'' is mainly aerobic. [[Glycolysis]] and the [[Tricarboxylic Acid (TCA) cycle]] are complete and functional in the bacteria. However, it is able to grow under anaerobic conditions in the presence of nitrate, though growth is much slower. | [[Cellular respiration]] in ''B. subtilis'' is mainly aerobic. [[Glycolysis]] and the [[Tricarboxylic Acid (TCA) cycle]] are complete and functional in the bacteria. However, it is able to grow under anaerobic conditions in the presence of nitrate, though growth is much slower. | ||
''B. subtilis'' has the ability to produce acid from a variety of sugars. In response to nutrient limitation, it also produces several [[enzymes]] (including proteases, amylases, cellulases, and lipases) and [[antibiotics]]. These enzymes are believed to help search for and degrade macromolecules to be used as energy sources, while the antibiotics are used to limit bacterial competition for the discovered energy sources. Furthermore, ''B. subtilis''contains 34 different two-component systems. [[ Two-component systems]] | ''B. subtilis'' has the ability to produce acid from a variety of sugars. In response to nutrient limitation, it also produces several [[enzymes]] (including proteases, amylases, cellulases, and lipases) and [[antibiotics]]. These enzymes are believed to help search for and degrade macromolecules to be used as energy sources, while the antibiotics are used to limit bacterial competition for the newly discovered energy sources. Furthermore, ''B. subtilis'' contains 34 different two-component systems. [[ Two-component systems]] consist of proteins that can detect environmental conditions and initiate the appropriate responses to these conditions. This large number of two-component systems suggests that ''B. subtilis'' can respond to a variety of environmental conditions effectively with a wide range of responses. <ref>Devine, Kevin M. (2000). "Bacillus subtilis: Genetics."</ref> | ||
''B. subtilis'' often exists as an [[endospore]]. As an endospore, ''B. subtilis'' becomes resistant to heat, acid, salt, and other harsh environmental conditions. Endospore formation is usually triggered by a lack of carbon, nitrogen or phosphate in the environment. This environmental "signal" is sensed by ''B. subtilis''' two-component systems, which initiate a [[phosphorelay]] by transferring a phosphate to Spo0A, one of the early transcriptional regulators involved in endospore formation. Hundreds of genes in the ''B. subtilis'' genome are involved in this phosphorelay. Asymmetric cell division proceeds, producing a large mother cell and a smaller, ellipse-shaped spore. The mother cell first engulfs the spore, and, following a series of morphological changes, the spore cortex and spore coat are then formed. The mother cell lyses and releases the newly formed endospore, which can survive through the poor environmental conditions.<ref>[http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=76 ''Bacillus subtilis Genome Project'']</ref><ref>Devine, Kevin M. (2000). "Bacillus subtilis: Genetics."</ref> | |||
[[Image: bsubtilisspore.jpg]] | [[Image: bsubtilisspore.jpg]] | ||
==Ecology== | ==Ecology== | ||
''B. subtilis'' is an important soil microorganism. It produces a variety of enzymes, which allow it to degrade many natural substrates and thus cycle nutrients in the soil. This increases nutrient availability to plants, thereby stimulating plant growth. Moreover, ''B. subtilis'' secretes [[antifungal]] antibiotics, which can control [[fungal diseases]] in plants and crops. ''B. subtilis'' is capable of increasing the overall performance of crops and other plants.<ref>{{cite journal|last=Malviya|first=M.K.|coauthors=A. Sharma, A. Pandey, Rinu K, P. Sati, and L.M.S. Palni|date=2012|title=''Bacillus subtilis'' NRRL B-30408: A potential inoculant for crops grown under rainfed conditions in the mountains|url=http://ref.scielo.org/hx53nb|journal=Journal of Soil Science and Plant Nutrition|publisher=Chilean Society of Soil Science|volume=12|issue=4|pages=811-824|issn=0718-9516|doi=10.4067/S0718-95162012005000034|accessdate=30 October 2013}}</ref> | |||
''B. subtilis'' is an important soil microorganism. It produces a variety of enzymes, which allow it to degrade many natural substrates and thus cycle nutrients in the soil. This increases nutrient availability to plants, thereby stimulating plant growth. Moreover, ''B. subtilis'' secretes [[antifungal]] antibiotics, which can control [[fungal diseases]] in plants and crops. ''B. subtilis'' is capable of increasing the overall performance of crops and other plants. | |||
==Pathology== | ==Pathology== | ||
''B. subtilis'' is not [[pathogenic]] to humans, animals, or plants. Although it may contaminate certain foods, such as cocoa and spices, it rarely causes food poisoning. ''B. subtilis'' does produce one extracellular [[toxin]], the enzyme [[subtilisin]]. Subtilisin can result in allergic reactions in some individuals; however, the allergic reaction will only occur following prolonged and repeated exposure. | ''B. subtilis'' is not [[pathogenic]] to humans, animals, or plants. Although it may contaminate certain foods, such as cocoa and spices, it rarely causes food poisoning. ''B. subtilis'' does produce one extracellular [[toxin]], the enzyme [[subtilisin]]. Subtilisin can result in allergic reactions in some individuals; however, the allergic reaction will only occur following prolonged and repeated exposure. | ||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
''B. subtilis'' is widely used in the field of [[biotechnology]] due to its ease at being manipulated and its generally low risk. ''B. subtilis'' produces a variety of enzymes useful to industry. The [[amylases]] it produces are used in the desizing of textiles and in starch modifications used to resize paper. The [[proteases]] it produces are used as additives in laundry detergents and products used for processing leather. ''B. subtilis'' also produces a wide array of [[antibiotics]], including subtilin, surfactin, bacillomycin, bacilysin, and fengycin, which are used as antibacterial and antifungal medicines. The antifungal activity of ''B. subtilis'' strain QST713 is used as a biological control agent of [[fungal plant pathogens]]. ''B. subtilis'' is also used as a [[soil inoculant]] in horticulture and agriculture. Furthermore, ''B. subtilis'' has been used to convert explosives into harmless compounds and to safely discard [[radionuclide wastes]]. ''B. subtilis'' was also used as a [[biowarfare simulant]] in [[Project SHAD]]. Project SHAD was a series of chemical and biological warfare tests conducted by the U.S. military during the Cold War era. Military personnel were exposed to biological and chemical agents, including ''B. subtilis'', to determine military vulnerabilities to such biowarfare agents and to determine appropriate response procedures.<ref>{{cite journal|last=Christopher|first=George W.|coauthors=Theodore J. Cieslak; Julie A. Pavlin; Edward M. Eitzen, Jr.|date=6 August 1997|title=Biological Warfare: A Historical Perspective|url=http://www2.cdc.gov/nip/isd/spoxclincian/contents/references/BioWarfareHistory.pdf|journal=Journal of the American Medical Association|volume=278|issue=5|pages=412-417|accessdate=30 October 2013}}</ref> | |||
''B. subtilis'' is also used directly in the food industry. A strain of ''B. subtilis'' is used in the production of [[natto]], a Japanese delicacy, and [[cheonggukjang]], a similar Korean delicacy. Natto is made by mixing soybeans with ''B. subtilis''. The mixture is [[fermented]] at 40 degrees Celsius for up to twenty-four hours. Subsequently, it is cooled and aged in a refrigerator for up to one week. This entire process creates the stringiness that is characteristic of natto.<ref>{{cite journal|last=Hesseltine|first=C. W.|date=October 1983|title=Microbiology of Oriental Fermented Foods|url=http://www.annualreviews.org/doi/abs/10.1146/annurev.mi.37.100183.003043|journal=Annual Review of Microbiology|volume=37|pages=575-601|id=6357061|doi=10.1146/annurev.mi.37.100183.003043|accessdate=30 October 2013}}</ref> | |||
''B. subtilis'' is also used directly in the food industry. A strain of ''B. subtilis'' is used in the production of [[natto]], a Japanese delicacy, and [[cheonggukjang]], a similar Korean delicacy. Natto is made by mixing soybeans with ''B. subtilis''. The mixture is [[fermented]] at 40 degrees | |||
==Current Research== | ==Current Research== | ||
"Induction of apoptosis in human leukemia K562 cells by cyclic lipopeptide from ''Bacillus subtilis'' natto T-2." (2007)<ref>[http://dx.doi.org/10.1016/j.peptides.2007.06.014 "Induction of apoptosis in human leukemia K562 cells by cyclic lipopeptide from Bacillus subtilis natto T-2."]</ref> | |||
"Induction of apoptosis in human leukemia K562 cells by cyclic lipopeptide from ''Bacillus subtilis'' natto T-2." (2007) | |||
Recent cancer research has focused much attention on inducing [[apoptosis]] of cancerous cells, as well as finding natural products with cancer-fighting effects. This research study examined the potential anti-cancer effects of a cyclic [[lipopeptide]] isolated from the natto strain of ''Bacillus subtilis''. This natto strain is safe to humans and can be found in a traditional Japanese food called "natto". The cyclic lipopeptide, in previous studies, exhibited anti-tumor properties. This study aimed to evaluate the cell growth-inhibiting and apoptosis-inducing effects of the cyclic lipopeptide from ''B. subtilis'' on human [[leukemia]] K562 cells. Researchers isolated the natto strain of ''B. subtilis'' directly from the Japanese food natto and obtained from it the cyclic lipopeptide. Leukemia K562 cells were treated with various concentrations of the cyclic lipopeptide, and [[fluorescent staining]], [[flow cytometric analysis]] and [[Western blotting]] were used for analysis. Fluorescent staining showed that the cyclic lipopeptide did induce apoptosis in the leukemia cells. Flow cytometric analysis indicated that the number of apoptotic cells increased with increasing concentrations of cyclic lipopeptide. Western blotting revealed that the apoptosis could be associated with the proteins caspase-3 and poly(ADP-ribose)polymerase. Therefore, this study provided evidence that lipopeptides from the natto strain of ''B. subtilis'' do have anti-tumor activity and can inhibit the proliferation of leukemia cells by inducing their apoptosis. With further research, this ''B. subtilis'' strain may prove to be an effective therapeutic agent against cancer. | Recent cancer research has focused much attention on inducing [[apoptosis]] of cancerous cells, as well as finding natural products with cancer-fighting effects. This research study examined the potential anti-cancer effects of a cyclic [[lipopeptide]] isolated from the natto strain of ''Bacillus subtilis''. This natto strain is safe to humans and can be found in a traditional Japanese food called "natto". The cyclic lipopeptide, in previous studies, exhibited anti-tumor properties. This study aimed to evaluate the cell growth-inhibiting and apoptosis-inducing effects of the cyclic lipopeptide from ''B. subtilis'' on human [[leukemia]] K562 cells. Researchers isolated the natto strain of ''B. subtilis'' directly from the Japanese food natto and obtained from it the cyclic lipopeptide. Leukemia K562 cells were treated with various concentrations of the cyclic lipopeptide, and [[fluorescent staining]], [[flow cytometric analysis]] and [[Western blotting]] were used for analysis. Fluorescent staining showed that the cyclic lipopeptide did induce apoptosis in the leukemia cells. Flow cytometric analysis indicated that the number of apoptotic cells increased with increasing concentrations of cyclic lipopeptide. Western blotting revealed that the apoptosis could be associated with the proteins caspase-3 and poly(ADP-ribose)polymerase. Therefore, this study provided evidence that lipopeptides from the natto strain of ''B. subtilis'' do have anti-tumor activity and can inhibit the proliferation of leukemia cells by inducing their apoptosis. With further research, this ''B. subtilis'' strain may prove to be an effective therapeutic agent against cancer. | ||
"Oral administration of a ''Bacillus subtilis'' spore-based vaccine expressing ''Clonorchis sinensis'' tegumental protein 22.3 kDa confers protection against ''Clonorchis sinensis''" (2008)<ref>[http://dx.doi.org/10.1016/j.vaccine.2008.02.015 "Oral administration of a Bacillus subtilis spore-based vaccine expressing Clonorchis sinensis tegumental protein 22.3 kDa confers protection against Clonorchis sinensis"]</ref> | |||
"Oral administration of a ''Bacillus subtilis'' spore-based vaccine expressing ''Clonorchis sinensis'' tegumental protein 22.3 kDa confers protection against ''Clonorchis sinensis''" (2008) | |||
[[Clonorchiasis]] is a bacterical disease caused by infection with the [[trematode]] ''Clonorchis sinensis''. It is found predominantly in Southern China, Korea, and other Southeast Asian countries and affects approximately 35 million people in the world. This study aimed to determine the effectiveness of an oral [[vaccine]], formed using the ''Bacillus subtilis'' endospore, against clonorchiasis. Due to its strong resistance to harsh environments, the ''B. subtilis'' endospore would provide an ideal transport system for the vaccine, which would need to penetrate the extreme environment of the [[gastrointestinal tract]]. Researchers thus used CotC, a major component of the ''B. subtilis'' [[spore coat]] and fused it with a novel [[tegumental protein gene]] from ''C. sinensis''. This tegumental protein was a membrane-bound protein with characteristics that made it a good vaccine candidate. [[Western blotting]] and [[immunofluorescence]] analyses were used to identify and confirm the tegumental protein expression on spores. Those recombinant spores that successfully displayed the tegumental protein were used for oral immunization. They were shown to generate an immunological response in rats infected with ''C. sinensis''. This study showed that oral administration of the [[recombinant]] spore can provide protection against C. sinensis. ''B. subtilis'' spores, thus, are a promising potential oral vaccine vehicle for the prevention and control of clonorchiasis and other gastrointestinal parasites. | [[Clonorchiasis]] is a bacterical disease caused by infection with the [[trematode]] ''Clonorchis sinensis''. It is found predominantly in Southern China, Korea, and other Southeast Asian countries and affects approximately 35 million people in the world. This study aimed to determine the effectiveness of an oral [[vaccine]], formed using the ''Bacillus subtilis'' endospore, against clonorchiasis. Due to its strong resistance to harsh environments, the ''B. subtilis'' endospore would provide an ideal transport system for the vaccine, which would need to penetrate the extreme environment of the [[gastrointestinal tract]]. Researchers thus used CotC, a major component of the ''B. subtilis'' [[spore coat]] and fused it with a novel [[tegumental protein gene]] from ''C. sinensis''. This tegumental protein was a membrane-bound protein with characteristics that made it a good vaccine candidate. [[Western blotting]] and [[immunofluorescence]] analyses were used to identify and confirm the tegumental protein expression on spores. Those recombinant spores that successfully displayed the tegumental protein were used for oral immunization. They were shown to generate an immunological response in rats infected with ''C. sinensis''. This study showed that oral administration of the [[recombinant]] spore can provide protection against C. sinensis. ''B. subtilis'' spores, thus, are a promising potential oral vaccine vehicle for the prevention and control of clonorchiasis and other gastrointestinal parasites. | ||
"Enhanced recombinant protein productivity by genome reduction in ''Bacillus subtilis''." (2008)<ref>[http://dx.doi.org/10.1093/dnares/dsn002 "Enhanced recombinant protein productivity by genome reduction in Bacillus subtilis."]</ref> | |||
"Enhanced recombinant protein productivity by genome reduction in ''Bacillus subtilis''." (2008) | |||
''Bacillus subtilis'' has been used extensively as a model microorganism. In this research study, ''B. subtilis'' was utilized as a [[model organism]] in the field of [[synthetic genomics]], which aims to facilitate the formation of microorganisms capable of enhanced productivity. Researchers created a new strain of ''B. subtilis'', MBG874, by depleting it of 874 kb (20%) of its normal genomic sequence. Deletions were completed in genes that did not affect cell growth and that were non-essential to cell function. This new strain's enzyme activity, specifically the activity of [[proteases]] and [[cellulases]], was then analyzed. Results revealed that, when compared with [[wild-type]] ''B. subtilis'', the productivity of extracellular cellulase and protease was enhanced. This study could not determine whether this enhanced enzyme production was due to a synergistic effect of genome reduction or due to the deletion of specific regulators. Further studies may facilitate the production of even more improved ''B. subtilis'' or other bacterial strains, which can be used in industry to produce optimal levels of specific proteins. | ''Bacillus subtilis'' has been used extensively as a model microorganism. In this research study, ''B. subtilis'' was utilized as a [[model organism]] in the field of [[synthetic genomics]], which aims to facilitate the formation of microorganisms capable of enhanced productivity. Researchers created a new strain of ''B. subtilis'', MBG874, by depleting it of 874 kb (20%) of its normal genomic sequence. Deletions were completed in genes that did not affect cell growth and that were non-essential to cell function. This new strain's enzyme activity, specifically the activity of [[proteases]] and [[cellulases]], was then analyzed. Results revealed that, when compared with [[wild-type]] ''B. subtilis'', the productivity of extracellular cellulase and protease was enhanced. This study could not determine whether this enhanced enzyme production was due to a synergistic effect of genome reduction or due to the deletion of specific regulators. Further studies may facilitate the production of even more improved ''B. subtilis'' or other bacterial strains, which can be used in industry to produce optimal levels of specific proteins. | ||
==References== | ==References== | ||
{{reflist|2}}[[Category:Suggestion Bot Tag]] | |||
[ | |||
[ | |||
Latest revision as of 16:00, 15 July 2024
 | ||||||||||||||
| Scientific classification | ||||||||||||||
| ||||||||||||||
| Binomial name | ||||||||||||||
| Bacillus subtilis |
Description and significance
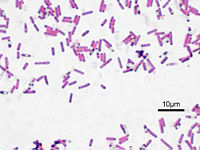
Bacillus subtilis is a Gram-positive, rod-shaped, endospore-forming bacteria. It is regarded as an aerobe; however, it is also capable of growing and sporulating under anaerobic conditions when necessary. B. subtilis resides predominantly in soil, including low-nutrient soil. Due to its association with soil particles, it is also inevitably transferred to plants, foods, animals and even marine and freshwater habitats.
B. subtilis was one of the first bacteria studied by scientists. It was originally named Vibrio subtilis in 1835 by Christian Gottfried Ehrenberg and later renamed Bacillus subtilis by Ferdinand Cohn in 1872.
Currently, B. subtilis is a significant microorganism in the scientific research field, as well as in biotechnology and industry. Scientists often utilize B. subtilis as a model organism. B. subtilis becomes naturally competent during its transition between exponential growth and the stationary phase of growth, meaning it is able to bend and internalize DNA from a medium. Thus, it is easily manipulated genetically and a good laboratory microorganism. Due to its endospore-forming mechanisms, it is an especially excellent model system for cell differentiation. Furthermore, B. subtilis produces a variety of enzymes used in biotechnology and industry, including antibiotic production. Its status as a "generally regarded as safe" (GRAS) organism makes it an excellent industrial microorganism.
Genome structure
The genome sequence of Bacillus subtilis was completed and published in November 1997.[1][2] It was the first Gram-positive bacteria to have its genome sequenced. B. subtilis contains a single, circular chromosome. It consists of 4,214,810 base pairs. The average G+C content is 43.5%, although ten regions have a G+C content significantly lower than average. There is a significant G-T skew at the third codon positions in common with other bacteria. Its leading strand has a 9% excess of G and 4% excess of T, as compared to the lagging strand. Approximately 87% of the genome consists of protein-coding regions. More than 74% of the open reading frames and 94% of ribosomal genes are transcribed co-directionally with replication. Only 53% of the genes are represented once. The remaining belong to multigene families, which range in size from 2 to 77 gene copies. Approximately 220 transcriptional regulators have been identified in the genome. At the present time, the functions of only approximately 58% of the genes are known, while the functions of 42% are still unknown and being studied. Twelve percent of these genes with unknown functions have homologs in other organisms.[3]
Replication of the B. subtilis genome proceeds similarly to most bacteria, with the splitting of the chromosome into two replication forks. B. subtilis has an origin of replication (oriC) and a terminus of replication (terC) located almost perfectly opposite each other on the genome. However, some studies have shown that the clockwise and counterclockwise replication forks actually differ in length by about 170 base pairs. Thus, once the clockwise replication fork reaches the terminus, its synthesis is terminated, while synthesis of the counterclockwise fork continues until it finally reaches the terminus. .[4]
Cell structure and metabolism
Bacillus subtilis is a prokaryotic cell, lacking membrane-bound organelles. It is enclosed by a cell wall, consisting of large amounts of peptidoglycan (Gram-positive). It is rod, or bacillus, shaped and usually found in colonies with other B. subtilis. B. subtilis is flagellated and thus motile.
Cellular respiration in B. subtilis is mainly aerobic. Glycolysis and the Tricarboxylic Acid (TCA) cycle are complete and functional in the bacteria. However, it is able to grow under anaerobic conditions in the presence of nitrate, though growth is much slower.
B. subtilis has the ability to produce acid from a variety of sugars. In response to nutrient limitation, it also produces several enzymes (including proteases, amylases, cellulases, and lipases) and antibiotics. These enzymes are believed to help search for and degrade macromolecules to be used as energy sources, while the antibiotics are used to limit bacterial competition for the newly discovered energy sources. Furthermore, B. subtilis contains 34 different two-component systems. Two-component systems consist of proteins that can detect environmental conditions and initiate the appropriate responses to these conditions. This large number of two-component systems suggests that B. subtilis can respond to a variety of environmental conditions effectively with a wide range of responses. [5]
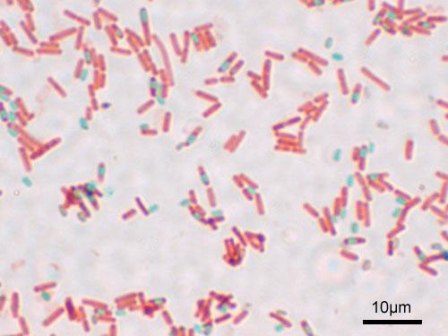
B. subtilis often exists as an endospore. As an endospore, B. subtilis becomes resistant to heat, acid, salt, and other harsh environmental conditions. Endospore formation is usually triggered by a lack of carbon, nitrogen or phosphate in the environment. This environmental "signal" is sensed by B. subtilis' two-component systems, which initiate a phosphorelay by transferring a phosphate to Spo0A, one of the early transcriptional regulators involved in endospore formation. Hundreds of genes in the B. subtilis genome are involved in this phosphorelay. Asymmetric cell division proceeds, producing a large mother cell and a smaller, ellipse-shaped spore. The mother cell first engulfs the spore, and, following a series of morphological changes, the spore cortex and spore coat are then formed. The mother cell lyses and releases the newly formed endospore, which can survive through the poor environmental conditions.[6][7]
Ecology
B. subtilis is an important soil microorganism. It produces a variety of enzymes, which allow it to degrade many natural substrates and thus cycle nutrients in the soil. This increases nutrient availability to plants, thereby stimulating plant growth. Moreover, B. subtilis secretes antifungal antibiotics, which can control fungal diseases in plants and crops. B. subtilis is capable of increasing the overall performance of crops and other plants.[8]
Pathology
B. subtilis is not pathogenic to humans, animals, or plants. Although it may contaminate certain foods, such as cocoa and spices, it rarely causes food poisoning. B. subtilis does produce one extracellular toxin, the enzyme subtilisin. Subtilisin can result in allergic reactions in some individuals; however, the allergic reaction will only occur following prolonged and repeated exposure.
Application to Biotechnology
B. subtilis is widely used in the field of biotechnology due to its ease at being manipulated and its generally low risk. B. subtilis produces a variety of enzymes useful to industry. The amylases it produces are used in the desizing of textiles and in starch modifications used to resize paper. The proteases it produces are used as additives in laundry detergents and products used for processing leather. B. subtilis also produces a wide array of antibiotics, including subtilin, surfactin, bacillomycin, bacilysin, and fengycin, which are used as antibacterial and antifungal medicines. The antifungal activity of B. subtilis strain QST713 is used as a biological control agent of fungal plant pathogens. B. subtilis is also used as a soil inoculant in horticulture and agriculture. Furthermore, B. subtilis has been used to convert explosives into harmless compounds and to safely discard radionuclide wastes. B. subtilis was also used as a biowarfare simulant in Project SHAD. Project SHAD was a series of chemical and biological warfare tests conducted by the U.S. military during the Cold War era. Military personnel were exposed to biological and chemical agents, including B. subtilis, to determine military vulnerabilities to such biowarfare agents and to determine appropriate response procedures.[9]
B. subtilis is also used directly in the food industry. A strain of B. subtilis is used in the production of natto, a Japanese delicacy, and cheonggukjang, a similar Korean delicacy. Natto is made by mixing soybeans with B. subtilis. The mixture is fermented at 40 degrees Celsius for up to twenty-four hours. Subsequently, it is cooled and aged in a refrigerator for up to one week. This entire process creates the stringiness that is characteristic of natto.[10]
Current Research
"Induction of apoptosis in human leukemia K562 cells by cyclic lipopeptide from Bacillus subtilis natto T-2." (2007)[11]
Recent cancer research has focused much attention on inducing apoptosis of cancerous cells, as well as finding natural products with cancer-fighting effects. This research study examined the potential anti-cancer effects of a cyclic lipopeptide isolated from the natto strain of Bacillus subtilis. This natto strain is safe to humans and can be found in a traditional Japanese food called "natto". The cyclic lipopeptide, in previous studies, exhibited anti-tumor properties. This study aimed to evaluate the cell growth-inhibiting and apoptosis-inducing effects of the cyclic lipopeptide from B. subtilis on human leukemia K562 cells. Researchers isolated the natto strain of B. subtilis directly from the Japanese food natto and obtained from it the cyclic lipopeptide. Leukemia K562 cells were treated with various concentrations of the cyclic lipopeptide, and fluorescent staining, flow cytometric analysis and Western blotting were used for analysis. Fluorescent staining showed that the cyclic lipopeptide did induce apoptosis in the leukemia cells. Flow cytometric analysis indicated that the number of apoptotic cells increased with increasing concentrations of cyclic lipopeptide. Western blotting revealed that the apoptosis could be associated with the proteins caspase-3 and poly(ADP-ribose)polymerase. Therefore, this study provided evidence that lipopeptides from the natto strain of B. subtilis do have anti-tumor activity and can inhibit the proliferation of leukemia cells by inducing their apoptosis. With further research, this B. subtilis strain may prove to be an effective therapeutic agent against cancer.
"Oral administration of a Bacillus subtilis spore-based vaccine expressing Clonorchis sinensis tegumental protein 22.3 kDa confers protection against Clonorchis sinensis" (2008)[12]
Clonorchiasis is a bacterical disease caused by infection with the trematode Clonorchis sinensis. It is found predominantly in Southern China, Korea, and other Southeast Asian countries and affects approximately 35 million people in the world. This study aimed to determine the effectiveness of an oral vaccine, formed using the Bacillus subtilis endospore, against clonorchiasis. Due to its strong resistance to harsh environments, the B. subtilis endospore would provide an ideal transport system for the vaccine, which would need to penetrate the extreme environment of the gastrointestinal tract. Researchers thus used CotC, a major component of the B. subtilis spore coat and fused it with a novel tegumental protein gene from C. sinensis. This tegumental protein was a membrane-bound protein with characteristics that made it a good vaccine candidate. Western blotting and immunofluorescence analyses were used to identify and confirm the tegumental protein expression on spores. Those recombinant spores that successfully displayed the tegumental protein were used for oral immunization. They were shown to generate an immunological response in rats infected with C. sinensis. This study showed that oral administration of the recombinant spore can provide protection against C. sinensis. B. subtilis spores, thus, are a promising potential oral vaccine vehicle for the prevention and control of clonorchiasis and other gastrointestinal parasites.
"Enhanced recombinant protein productivity by genome reduction in Bacillus subtilis." (2008)[13]
Bacillus subtilis has been used extensively as a model microorganism. In this research study, B. subtilis was utilized as a model organism in the field of synthetic genomics, which aims to facilitate the formation of microorganisms capable of enhanced productivity. Researchers created a new strain of B. subtilis, MBG874, by depleting it of 874 kb (20%) of its normal genomic sequence. Deletions were completed in genes that did not affect cell growth and that were non-essential to cell function. This new strain's enzyme activity, specifically the activity of proteases and cellulases, was then analyzed. Results revealed that, when compared with wild-type B. subtilis, the productivity of extracellular cellulase and protease was enhanced. This study could not determine whether this enhanced enzyme production was due to a synergistic effect of genome reduction or due to the deletion of specific regulators. Further studies may facilitate the production of even more improved B. subtilis or other bacterial strains, which can be used in industry to produce optimal levels of specific proteins.
References
- ↑ | B. subtilis Genome
- ↑ "The complete genome sequence of the Gram-positive bacterium Bacillus subtilis."
- ↑ Devine, Kevin M. (2000). "Bacillus subtilis: Genetics."
- ↑ Lonenshein, A.L., Hoch, J.A., & Losick, R. (1993). Bacillus subtilis and other Gram-positive bacteria.
- ↑ Devine, Kevin M. (2000). "Bacillus subtilis: Genetics."
- ↑ Bacillus subtilis Genome Project
- ↑ Devine, Kevin M. (2000). "Bacillus subtilis: Genetics."
- ↑ Malviya, M.K.; A. Sharma, A. Pandey, Rinu K, P. Sati, and L.M.S. Palni (2012). "Bacillus subtilis NRRL B-30408: A potential inoculant for crops grown under rainfed conditions in the mountains". Journal of Soil Science and Plant Nutrition 12 (4): 811-824. DOI:10.4067/S0718-95162012005000034. ISSN 0718-9516. Retrieved on 30 October 2013. Research Blogging.
- ↑ Christopher, George W.; Theodore J. Cieslak; Julie A. Pavlin; Edward M. Eitzen, Jr. (6 August 1997). "Biological Warfare: A Historical Perspective". Journal of the American Medical Association 278 (5): 412-417. Retrieved on 30 October 2013.
- ↑ Hesseltine, C. W. (October 1983). "Microbiology of Oriental Fermented Foods". Annual Review of Microbiology 37: 575-601. DOI:10.1146/annurev.mi.37.100183.003043. 6357061. Retrieved on 30 October 2013. Research Blogging.
- ↑ "Induction of apoptosis in human leukemia K562 cells by cyclic lipopeptide from Bacillus subtilis natto T-2."
- ↑ "Oral administration of a Bacillus subtilis spore-based vaccine expressing Clonorchis sinensis tegumental protein 22.3 kDa confers protection against Clonorchis sinensis"
- ↑ "Enhanced recombinant protein productivity by genome reduction in Bacillus subtilis."