Evolution of cells: Difference between revisions
imported>David Tribe |
mNo edit summary |
||
| (48 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
{{ | {{subpages}} | ||
<blockquote>"The evolution of modern cells is arguably the most challenging and important problem the field of Biology has ever faced. In Darwin's day the problem could hardly be imagined. For much of the 20th century it was intractable. In any case, the problem lay buried in the catch-all rubric "origin of life"---where, because it is a biological not a (bio)chemical problem, it was effectively ignored. Scientific interest in cellular evolution started to pick up once the universal phylogenetic tree, the framework within which the problem had to be addressed, was determined . But it was not until microbial genomics arrived on the scene that biologists could actually do much about the problem of cellular evolution." ([[Carl Woese]], 2002) <ref> Woese C (2002) [http://www.pnas.org/cgi/content/full/99/13/8742 On the evolution of cells.] ''Proc Natl Acad Sci USA'' '''99''':8742-7 PMID 12077305 This article shifts the emphasis in early [[Phylogenetics|phylogenic adaptation]] from vertical to horizontal gene transfer. (Open access.)</ref></blockquote> | <blockquote>"The evolution of modern cells is arguably the most challenging and important problem the field of Biology has ever faced. In Darwin's day the problem could hardly be imagined. For much of the 20th century it was intractable. In any case, the problem lay buried in the catch-all rubric "origin of life"---where, because it is a biological not a (bio)chemical problem, it was effectively ignored. Scientific interest in cellular evolution started to pick up once the universal phylogenetic tree, the framework within which the problem had to be addressed, was determined . But it was not until microbial genomics arrived on the scene that biologists could actually do much about the problem of cellular evolution." ([[Carl Woese]], 2002) <ref> Woese C (2002) [http://www.pnas.org/cgi/content/full/99/13/8742 On the evolution of cells.] ''Proc Natl Acad Sci USA'' '''99''':8742-7 PMID 12077305 This article shifts the emphasis in early [[Phylogenetics|phylogenic adaptation]] from vertical to horizontal gene transfer. (Open access.)</ref></blockquote> | ||
<blockquote> | |||
The First Cell arose in the previously pre-biotic world with the coming together | |||
of several entities that gave a single vesicle the unique chance to carry out | |||
three essential and quite different life processes. These were: (a) to copy | |||
informational macromolecules, (b) to carry out specific catalytic functions, and | |||
(c) to couple energy from the environment into usable chemical forms. These | |||
would foster subsequent cellular evolution and metabolism. Each of these three | |||
essential processes probably originated and was lost many times prior to The | |||
First Cell, but only when these three occurred together was life jump-started | |||
and Darwinian evolution of organisms began. (Koch and Silver, 2005)<ref>Koch AL, Silver S.(2005) The first cell. Adv Microb Physiol. 50:227-59. PMID 16221582</ref> </blockquote> | |||
==The first cells== | ==The first cells== | ||
The origin of cells was the most important step in the evolution of life as we know it. The birth of the cell marked the passage from | The origin of cells was the most important step in the evolution of life as we know it. The birth of the cell marked the passage from pre-biotic chemistry to partitioned units resembling modern cells. The final transition to living entities that fulfill all the definitions of modern cells depended on the ability to evolve effectively by natural selection. This transition has been called the [[Darwinian transition]]. | ||
If life is viewed from the point of view of [[replicator]] molecules, cells satisfy two fundamental conditions: protection from the outside environment and confinement of biochemical activity. The former condition is needed to keep complex molecules stable in a varying and sometimes aggressive environment; the latter is fundamental for the evolution of [[biological complexity]]. If freely-floating molecules that code for [[enzyme]]s are not enclosed in cells, the enzymes will automatically benefit the neighbouring replicator molecules. The consequences of diffusion in non-partitioned life forms might be viewed as "[[parasitism]] by default." Therefore the [[natural selection|selection pressure]] on replicator molecules will be lower, as the 'lucky' molecule that produces the better enzyme has no definitive advantage over its close neighbors. If the molecule is enclosed in a cell membrane, then the enzymes coded will be available only to the replicator molecule itself. That molecule will uniquely benefit from the enzymes it codes for; giving it a better chance to multiply. | If life is viewed from the point of view of [[replicator]] molecules, cells satisfy two fundamental conditions: protection from the outside environment and confinement of biochemical activity. The former condition is needed to keep complex molecules stable in a varying and sometimes aggressive environment; the latter is fundamental for the evolution of [[biological complexity]]. If freely-floating molecules that code for [[enzyme]]s are not enclosed in cells, the enzymes will automatically benefit the neighbouring replicator molecules. The consequences of diffusion in non-partitioned life forms might be viewed as "[[parasitism]] by default." Therefore the [[natural selection|selection pressure]] on replicator molecules will be lower, as the 'lucky' molecule that produces the better enzyme has no definitive advantage over its close neighbors. If the molecule is enclosed in a cell membrane, then the enzymes coded will be available only to the replicator molecule itself. That molecule will uniquely benefit from the enzymes it codes for; giving it a better chance to multiply. | ||
| Line 12: | Line 25: | ||
Another theory holds that the turbulent shores of the ancient coastal waters may have served as a mammoth laboratory, aiding in the countless experiments necessary to bring about the first cell. Waves breaking on the shore create a delicate foam composed of bubbles. Winds sweeping across the ocean have a tendency to drive things to shore, much like driftwood collecting on the beach. It is possible that organic molecules were concentrated on the shorelines in much the same way. Shallow coastal waters also tend to be warmer, further concentrating the molecules through [[evaporation]]. While bubbles comprised of mostly water tend to burst quickly, oily bubbles happen to be much more stable, lending more time to the particular bubble to perform these crucial experiments. The [[Phospholipid|phospholipid]] is a good example of a common oily compound prevalent in the prebiotic seas. | Another theory holds that the turbulent shores of the ancient coastal waters may have served as a mammoth laboratory, aiding in the countless experiments necessary to bring about the first cell. Waves breaking on the shore create a delicate foam composed of bubbles. Winds sweeping across the ocean have a tendency to drive things to shore, much like driftwood collecting on the beach. It is possible that organic molecules were concentrated on the shorelines in much the same way. Shallow coastal waters also tend to be warmer, further concentrating the molecules through [[evaporation]]. While bubbles comprised of mostly water tend to burst quickly, oily bubbles happen to be much more stable, lending more time to the particular bubble to perform these crucial experiments. The [[Phospholipid|phospholipid]] is a good example of a common oily compound prevalent in the prebiotic seas. | ||
Phospholipids can be visualized in one's mind as a [[hydrophilic]] head on one end, and a [[hydrophobic]] tail on the other. These molecules possess an important characteristic for the construction of cell membranes; they are able to link together to form a [[Lipid bilayer|bilayer]] membrane. A lipid monolayer bubble can only contain oil, and is therefore not conducive to harbouring water-soluble organic molecules. On the other hand, a lipid bilayer bubble [http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/P/Phospholipids.html] can contain water, and was a likely precursor to the modern cell membrane. If a protein came along that increased the integrity of its parent bubble, then that bubble had an advantage, and was placed at the top of the [[natural selection]] waiting list. Primitive reproduction can be envisioned when the bubbles burst, releasing the results of the experiment into the surrounding medium. Once enough of the 'right stuff' was released into the medium, the development of the first [[prokaryotes]], [[eukaryotes]], and multi-cellular organisms could be achieved. <ref>This theory is expanded upon in ''The Cell: Evolution of the First Organism'' by [[Joseph Panno]]</ref> | Phospholipids can be visualized in one's mind as a [[hydrophilic]] head on one end, and a [[hydrophobic]] tail on the other. These molecules possess an important characteristic for the construction of cell membranes; they are able to link together to form a [[Lipid bilayer|bilayer]] membrane. A lipid monolayer bubble can only contain oil, and is therefore not conducive to harbouring water-soluble organic molecules. On the other hand, a lipid bilayer bubble [http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/P/Phospholipids.html] can contain water, and was a likely precursor to the modern cell membrane. If a protein came along that increased the integrity of its parent bubble, then that bubble had an advantage, and was placed at the top of the [[natural selection]] waiting list. Primitive reproduction can be envisioned when the bubbles burst, releasing the results of the experiment into the surrounding medium. Once enough of the 'right stuff' was released into the medium, the development of the first [[prokaryotes]], [[eukaryotes]], and multi-cellular organisms could be achieved. <ref>This theory is expanded upon in ''The Cell: Evolution of the First Organism'' by [[Joseph Panno]]</ref> | ||
==Community metabolism== | ==Community metabolism== | ||
The common ancestor of eukaryotes, bacteria, and archaea may well have been a community of organisms that readily exchanged components and genes. It would have contained: | The common ancestor of the now existing cellular lineages (eukaryotes, bacteria, and archaea) may well have been a community of organisms that readily exchanged components and genes. It would have contained: | ||
* [[Autotroph]]s that produced organic compounds from CO2 either photosynthetically or by inorganic chemical reactions; | * [[Autotroph]]s that produced organic compounds from CO2 either photosynthetically or by inorganic chemical reactions; | ||
* [[Heterotroph]]s that obtained organics by leakage from other organisms | * [[Heterotroph]]s that obtained organics by leakage from other organisms | ||
* [[Saprotrophs]] that absorbed nutrients from decaying organisms | * [[Saprotrophs]] that absorbed nutrients from decaying organisms | ||
* [[Phagotrophs]] that were sufficiently complex to envelop and digest particulate nutrients including other organisms | * [[Phagotrophs]] that were sufficiently complex to envelop and digest particulate nutrients including other organisms. | ||
The eukaryotic cell seems to have evolved from a [[symbiosis|symbiotic community]] of prokaryotic cells. It appears that DNA-bearing organelles like mitochondria and chloroplasts are remnants of ancient symbiotic oxygen-breathing [[bacterium|bacteria]] and [[cyanobacteria]], respectively, where at least part of the rest of the cell may have been derived from an ancestral [[archaea]]n prokaryote cell | The eukaryotic cell seems to have evolved from a [[symbiosis|symbiotic community]] of prokaryotic cells. It appears that DNA-bearing organelles like mitochondria and chloroplasts are remnants of ancient symbiotic oxygen-breathing [[bacterium|bacteria]] and [[cyanobacteria]], respectively, where at least part of the rest of the cell may have been derived from an ancestral [[archaea]]n prokaryote cell. This concept is often termed the [[endosymbiotic theory]] but is perhaps better considered as an [[hypothesis]]. There is still debate about whether organelles like the [[hydrogenosome]] predated the origin of [[mitochondria]], or ''vice versa'': see the [[hydrogen hypothesis]] for the origin of eukaryotic cells. | ||
How the current lineages of microbes evolved from this postulated community is currently unsolved but subject to intense research by biologists, stimulated by the great flow of new discoveries emerging from [[genomics|genome science]].<ref>Kurland CG ''et al.'' (2006) Genomics and the irreducible nature of eukaryote cells. ''Science'' '''312'''(5776):1011-4 PMID 16709776</ref> | How the current lineages of microbes evolved from this postulated community is currently unsolved but subject to intense research by biologists, stimulated by the great flow of new discoveries emerging from [[genomics|genome science]].<ref>Kurland CG ''et al.'' (2006) Genomics and the irreducible nature of eukaryote cells. ''Science'' '''312'''(5776):1011-4 PMID 16709776</ref> | ||
| Line 27: | Line 40: | ||
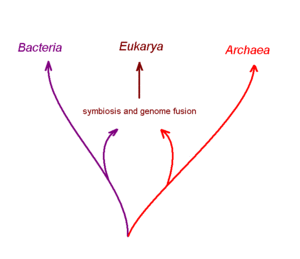
[[Image:Genome_fusion_eukarya.gif|thumb|300px|Right|One current hypothesis for the origin of eukaryotic (nucleated) cells, from fusion of two distinct non-nucleated prokaryotes that were partners of a postulated ancient [[symbiosis]].]] | [[Image:Genome_fusion_eukarya.gif|thumb|300px|Right|One current hypothesis for the origin of eukaryotic (nucleated) cells, from fusion of two distinct non-nucleated prokaryotes that were partners of a postulated ancient [[symbiosis]].]] | ||
== | ==Genetic code and the RNA world== | ||
Much modern evidence supports the hypothesis that early cellular evolution occurred in a biological realm radically distinct from modern biology. It is thought that in this ancient realm, the current genetic role of DNA was largely filled by RNA, and catalysis also was largely mediated by RNA (that is, by [[ribozyme]] counterparts of enzymes). This remarkable concept is known as the [[RNA world hypothesis]]. | |||
According to this hypothesis, the ancient RNA world transitioned into the modern cellular world via the evolution of forms of protein synthesis akin to modern ribosomal protein synthesis, followed by replacement of many cellular ribozyme catalysts by protein based enzyme catalysts. Proteins are much more flexible in catalysis that RNA due to the existence of diverse amino acid side chains with distinct chemical characteristics. The RNA record in existing cells appears to preserve some 'molecular fossils' from this RNA world. These RNA fossils include the ribosome itself (in which RNA catalyses peptide-bond formation), the modern ribozyme catalyst RNase P, and tRNAs. | |||
The universal [[Genetic code]] may indeed preserve some evidence for this ancient world. For instance, recent studies of transfer RNAs , the enzymes that charge them with amino acids (the first step in protein synthesis) and the way these components recognise and exploit the genetic code, have been used to suggest that of the universal genetic code emerged before the evolution of the modern amino acid activation method for protein synthesis.<ref> Hohn, M J 'et al.' (2006). Emergence of the universal genetic code imprinted in an RNA record. Proc. Natl. Acad. Sci. USA 103:18095-18100 PMID 17110438 | |||
* O'Donoghue P Luthey-Schulten Z (2003) [http://mmbr.asm.org/cgi/content/full/67/4/550?view=long&pmid=14665676 On the evolution of structure in aminoacyl-tRNA synthetases.] Microbiol Mol Biol Rev 67:550–573. PMID 14665676 | |||
* Jeffares DC Poole AM Penny D (1998) Relics from the RNA world. J Mol Evol. 46:18-36. Review. PMID 9419222 | |||
* Poole AM Jeffares DC Penny D (1998) The path from the RNA world. J Mol Evol. 46:1-17. Review. PMID 9419221 | |||
* Gesteland, RF ''et al.'' eds.(2006) The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA (2006) (Cold Spring Harbor Lab Press, Cold Spring Harbor, NY,).</ref> | |||
==Canonical patterns== | |||
Despite the fact that the evolutionary origins of the major lineages of modern cells are currently hotly disputed, the primary distinctions between the three major lineages of cellular life (called domains) are clear cut and firmly established. | Despite the fact that the evolutionary origins of the major lineages of modern cells are currently hotly disputed, the primary distinctions between the three major lineages of cellular life (called domains) are clear cut and firmly established. | ||
In each of these three domains, protein synthesis machinery, DNA replication, and transcription of RNA from DNA all display distinctive features. There are three versions of ribosomal RNAs, and three versions of each ribosomal protein, one for each domain of life.<ref> | In each of these three domains, protein synthesis machinery, DNA replication, and transcription of RNA from DNA all display distinctive features. There are three versions of ribosomal RNAs, and generally three versions of each ribosomal protein, one for each domain of life. These three versions of the protein synthesis apparatus are called the ''canonical patterns'', and it is the existence of these canonical patterns provide the basis for a definition of the three Domains - ''[[Bacteria]]'', ''[[Archaea]]'', and ''[[Eukaryote|Eukarya, (alternatively called Eukaryota)]]'' - of currently existing cells.<ref> | ||
Olsen G J Woese C R (1997) Archaeal genomics: an overview. Cell 89:991–994 PMID 9215619 | Olsen G J Woese C R (1997) Archaeal genomics: an overview. Cell 89:991–994 PMID 9215619 | ||
* Makarova K S Koonin E V (2005) Evolutionary and functional genomics of the Archaea. Curr. Opin. Microbiol 8 | * Makarova K S Koonin E V (2005) Evolutionary and functional genomics of the Archaea. Curr. Opin. Microbiol 8:586–594. PMID 16111915 | ||
* Woese C R 'et al.' (2000) [ http://mmbr.asm.org/cgi/content/abstract/64/1/202?ijkey=9104f15d5ac0a9f9f1cf4451691dc8343169a880&keytype2=tf_ipsecsha Aminoacyl-tRNA Synthetases, the Genetic Code, and the Evolutionary Process] Microbiol. Mol. Biol. Rev 64 | * Woese C R 'et al.' (2000) [http://mmbr.asm.org/cgi/content/abstract/64/1/202?ijkey=9104f15d5ac0a9f9f1cf4451691dc8343169a880&keytype2=tf_ipsecsha Aminoacyl-tRNA Synthetases, the Genetic Code, and the Evolutionary Process] Microbiol. Mol. Biol. Rev 64:202–236 | ||
* Forterre P (2006) Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: A hypothesis for the origin of cellular domain. PNAS 103:3669-3674</ref> | * Forterre P (2006) Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: A hypothesis for the origin of cellular domain. PNAS 103:3669-3674</ref> | ||
==Using genomics to infer early lines of evolution== | |||
Instead of relying a single gene such as the small-subunit ribosomal RNA (SSU rRNA) gene to reconstruct early evolution, or a few genes, scientific effort has shifted to exploiting the comprehensive information from the many complete genome sequences of organisms that are now available.<ref>Daubin V ''et al.'' (2003) Phylogenetics and the cohesion of bacterial genomes. ''Science'' '''301''':829-32 PMID 12907801 | Instead of relying a single gene such as the small-subunit ribosomal RNA (SSU rRNA) gene to reconstruct early evolution, or a few genes, scientific effort has shifted to exploiting the comprehensive information from the many complete genome sequences of organisms that are now available.<ref>Daubin V ''et al.'' (2003) Phylogenetics and the cohesion of bacterial genomes. ''Science'' '''301''':829-32 PMID 12907801 | ||
| Line 41: | Line 68: | ||
* Henz SR ''et al.'' (2005) Whole-genome prokaryotic phylogeny. ''Bioinformatics'' [http://bioinformatics.oxfordjournals.org/cgi/content/full/21/10/2329 '''21''':2329-35] PMID 15166018</ref> | * Henz SR ''et al.'' (2005) Whole-genome prokaryotic phylogeny. ''Bioinformatics'' [http://bioinformatics.oxfordjournals.org/cgi/content/full/21/10/2329 '''21''':2329-35] PMID 15166018</ref> | ||
From such studies, it is now clear that trees based only on SSU rRNA alone do not capture the events of early eukaryote evolution accurately, and the progenitors of the first nucleated cells are still uncertain. For instance, careful analysis of the complete genome of the eukaryote yeast shows that many of its genes are more closely related to bacterial genes than they are to archaea, and it is now clear that archaea were not the simple progenitors of the eukaryotes, in stark contradiction to the earlier findings based on SSU rRNA and limited samples of other genes.<ref>Esser C ''et al.'' (2004) A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. ''Mol Biol Evol'' '''21''':1643-50 PMID 15155797 | From such studies, it is now clear that evolututionary trees based only on SSU rRNA alone do not capture the events of early eukaryote evolution accurately, and the progenitors of the first nucleated cells are still uncertain. For instance, careful analysis of the complete genome of the eukaryote yeast shows that many of its genes are more closely related to bacterial genes than they are to archaea, and it is now clear that archaea were not the simple progenitors of the eukaryotes, in stark contradiction to the earlier findings based on SSU rRNA and limited samples of other genes.<ref>Esser C ''et al.'' (2004) A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. ''Mol Biol Evol'' '''21''':1643-50 PMID 15155797 | ||
*Rivera MC, Lake JA (2004) The ring of life provides evidence for a genome fusion origin of eukaryotes. ''Nature'' '''431''':152-5 PMID 15356622 | *Rivera MC, Lake JA (2004) The ring of life provides evidence for a genome fusion origin of eukaryotes. ''Nature'' '''431''':152-5 PMID 15356622 | ||
* Simonson AB ''et al.'' (2005) Decoding the genomic tree of life. ''Proc Natl Acad Sci USA'' '''102''' Suppl 1:6608-13 PMID 15851667</ref> | * Simonson AB ''et al.'' (2005) Decoding the genomic tree of life. ''Proc Natl Acad Sci USA'' '''102''' Suppl 1:6608-13 PMID 15851667</ref> | ||
One imaginative but disputed hypothesis getting some support from recent computer-assisted studies of complete genome DNA sequences is that the first nucleated cell arose from two distinctly different ancient prokaryotic (non-nucleated) species that had formed a [[symbiosis|symbiotic]] relationship with one-another to carry out different aspects of metabolism. One partner of this postulated symbiosis is proposed to be a true bacterial cell, and the other an archaean cell. It is postulated that this symbiotic partnership progressed via the cellular fusion of the partners to generate a [[chimera|chimeric]] or hybrid cell with a membrane bound internal structure that was the forerunner of the nucleus. The next stage in this scheme was transfer of both partner genomes into the nucleus and their fusion with one-another. Several variations of this hypothesis for the origin of nucleated cells have been suggested<ref>Esser C ''et al.'' (2004) A genome phylogeny for mitochondria among alpha-proteobacteria and a preedominantly eubacterial ancestry of yeast nuclear genes. ''Mol Biol Evol'' '''21''':1643-50 PMID 15155797</ref> | One imaginative but disputed hypothesis getting some support from recent computer-assisted studies of complete genome DNA sequences is that the first nucleated cell arose from two distinctly different ancient prokaryotic (non-nucleated) species that had formed a [[symbiosis|symbiotic]] relationship with one-another to carry out different aspects of metabolism. One partner of this postulated symbiosis is proposed to be a true bacterial cell, and the other an archaean cell. It is postulated that this symbiotic partnership progressed via the cellular fusion of the partners to generate a [[chimera|chimeric]] or hybrid cell with a membrane bound internal structure that was the forerunner of the nucleus. The next stage in this scheme was transfer of both partner genomes into the nucleus and their fusion with one-another. Several variations of this hypothesis for the origin of nucleated cells have been suggested.<ref>Esser C ''et al.'' (2004) A genome phylogeny for mitochondria among alpha-proteobacteria and a preedominantly eubacterial ancestry of yeast nuclear genes. ''Mol Biol Evol'' '''21''':1643-50 PMID 15155797</ref> | ||
But some biologists dispute this conception<ref>Kurland CG ''et al.'' (2006) Genomics and the irreducible nature of eukaryote cells. ''Science'' '''312'''(5776):1011-4 PMID 16709776</ref> and argue for the need for a shift in conceptual framework if this problem is to be solved - | But some biologists dispute this conception<ref>Kurland CG ''et al.'' (2006) Genomics and the irreducible nature of eukaryote cells. ''Science'' '''312'''(5776):1011-4 PMID 16709776</ref> and argue for the need for a shift in conceptual framework if this problem is to be solved. They re-iterate the community metabolism theme, the idea that early living communities would comprise many different entities to extant cells, and would have shared their genetic material more extensively than current microbes.<ref> Woese C (2002) [http://www.pnas.org/cgi/content/full/99/13/8742 On the evolution of cells.] ''Proc Natl Acad Sci USA'' '''99''':8742-7 PMID 12077305 </ref> | ||
== | ==References== | ||
<references/>[[Category:Suggestion Bot Tag]] | |||
[[Category: | |||
Latest revision as of 11:00, 14 August 2024
"The evolution of modern cells is arguably the most challenging and important problem the field of Biology has ever faced. In Darwin's day the problem could hardly be imagined. For much of the 20th century it was intractable. In any case, the problem lay buried in the catch-all rubric "origin of life"---where, because it is a biological not a (bio)chemical problem, it was effectively ignored. Scientific interest in cellular evolution started to pick up once the universal phylogenetic tree, the framework within which the problem had to be addressed, was determined . But it was not until microbial genomics arrived on the scene that biologists could actually do much about the problem of cellular evolution." (Carl Woese, 2002) [1]
The First Cell arose in the previously pre-biotic world with the coming together of several entities that gave a single vesicle the unique chance to carry out three essential and quite different life processes. These were: (a) to copy informational macromolecules, (b) to carry out specific catalytic functions, and (c) to couple energy from the environment into usable chemical forms. These would foster subsequent cellular evolution and metabolism. Each of these three essential processes probably originated and was lost many times prior to The First Cell, but only when these three occurred together was life jump-started
and Darwinian evolution of organisms began. (Koch and Silver, 2005)[2]
The first cells
The origin of cells was the most important step in the evolution of life as we know it. The birth of the cell marked the passage from pre-biotic chemistry to partitioned units resembling modern cells. The final transition to living entities that fulfill all the definitions of modern cells depended on the ability to evolve effectively by natural selection. This transition has been called the Darwinian transition.
If life is viewed from the point of view of replicator molecules, cells satisfy two fundamental conditions: protection from the outside environment and confinement of biochemical activity. The former condition is needed to keep complex molecules stable in a varying and sometimes aggressive environment; the latter is fundamental for the evolution of biological complexity. If freely-floating molecules that code for enzymes are not enclosed in cells, the enzymes will automatically benefit the neighbouring replicator molecules. The consequences of diffusion in non-partitioned life forms might be viewed as "parasitism by default." Therefore the selection pressure on replicator molecules will be lower, as the 'lucky' molecule that produces the better enzyme has no definitive advantage over its close neighbors. If the molecule is enclosed in a cell membrane, then the enzymes coded will be available only to the replicator molecule itself. That molecule will uniquely benefit from the enzymes it codes for; giving it a better chance to multiply.
How did partitioning begin? Biochemically, cell-like spheroids formed by proteinoids are observed by heating amino acids with phosphoric acid as a catalyst. They bear much of the basic features provided by cell membranes. Proteinoid-based protocells enclosing RNA molecules could (but not necessarily should) have been the first cellular life forms on Earth.
Another theory holds that the turbulent shores of the ancient coastal waters may have served as a mammoth laboratory, aiding in the countless experiments necessary to bring about the first cell. Waves breaking on the shore create a delicate foam composed of bubbles. Winds sweeping across the ocean have a tendency to drive things to shore, much like driftwood collecting on the beach. It is possible that organic molecules were concentrated on the shorelines in much the same way. Shallow coastal waters also tend to be warmer, further concentrating the molecules through evaporation. While bubbles comprised of mostly water tend to burst quickly, oily bubbles happen to be much more stable, lending more time to the particular bubble to perform these crucial experiments. The phospholipid is a good example of a common oily compound prevalent in the prebiotic seas.
Phospholipids can be visualized in one's mind as a hydrophilic head on one end, and a hydrophobic tail on the other. These molecules possess an important characteristic for the construction of cell membranes; they are able to link together to form a bilayer membrane. A lipid monolayer bubble can only contain oil, and is therefore not conducive to harbouring water-soluble organic molecules. On the other hand, a lipid bilayer bubble [1] can contain water, and was a likely precursor to the modern cell membrane. If a protein came along that increased the integrity of its parent bubble, then that bubble had an advantage, and was placed at the top of the natural selection waiting list. Primitive reproduction can be envisioned when the bubbles burst, releasing the results of the experiment into the surrounding medium. Once enough of the 'right stuff' was released into the medium, the development of the first prokaryotes, eukaryotes, and multi-cellular organisms could be achieved. [3]
Community metabolism
The common ancestor of the now existing cellular lineages (eukaryotes, bacteria, and archaea) may well have been a community of organisms that readily exchanged components and genes. It would have contained:
- Autotrophs that produced organic compounds from CO2 either photosynthetically or by inorganic chemical reactions;
- Heterotrophs that obtained organics by leakage from other organisms
- Saprotrophs that absorbed nutrients from decaying organisms
- Phagotrophs that were sufficiently complex to envelop and digest particulate nutrients including other organisms.
The eukaryotic cell seems to have evolved from a symbiotic community of prokaryotic cells. It appears that DNA-bearing organelles like mitochondria and chloroplasts are remnants of ancient symbiotic oxygen-breathing bacteria and cyanobacteria, respectively, where at least part of the rest of the cell may have been derived from an ancestral archaean prokaryote cell. This concept is often termed the endosymbiotic theory but is perhaps better considered as an hypothesis. There is still debate about whether organelles like the hydrogenosome predated the origin of mitochondria, or vice versa: see the hydrogen hypothesis for the origin of eukaryotic cells.
How the current lineages of microbes evolved from this postulated community is currently unsolved but subject to intense research by biologists, stimulated by the great flow of new discoveries emerging from genome science.[4]
Genetic code and the RNA world
Much modern evidence supports the hypothesis that early cellular evolution occurred in a biological realm radically distinct from modern biology. It is thought that in this ancient realm, the current genetic role of DNA was largely filled by RNA, and catalysis also was largely mediated by RNA (that is, by ribozyme counterparts of enzymes). This remarkable concept is known as the RNA world hypothesis.
According to this hypothesis, the ancient RNA world transitioned into the modern cellular world via the evolution of forms of protein synthesis akin to modern ribosomal protein synthesis, followed by replacement of many cellular ribozyme catalysts by protein based enzyme catalysts. Proteins are much more flexible in catalysis that RNA due to the existence of diverse amino acid side chains with distinct chemical characteristics. The RNA record in existing cells appears to preserve some 'molecular fossils' from this RNA world. These RNA fossils include the ribosome itself (in which RNA catalyses peptide-bond formation), the modern ribozyme catalyst RNase P, and tRNAs.
The universal Genetic code may indeed preserve some evidence for this ancient world. For instance, recent studies of transfer RNAs , the enzymes that charge them with amino acids (the first step in protein synthesis) and the way these components recognise and exploit the genetic code, have been used to suggest that of the universal genetic code emerged before the evolution of the modern amino acid activation method for protein synthesis.[5]
Canonical patterns
Despite the fact that the evolutionary origins of the major lineages of modern cells are currently hotly disputed, the primary distinctions between the three major lineages of cellular life (called domains) are clear cut and firmly established.
In each of these three domains, protein synthesis machinery, DNA replication, and transcription of RNA from DNA all display distinctive features. There are three versions of ribosomal RNAs, and generally three versions of each ribosomal protein, one for each domain of life. These three versions of the protein synthesis apparatus are called the canonical patterns, and it is the existence of these canonical patterns provide the basis for a definition of the three Domains - Bacteria, Archaea, and Eukarya, (alternatively called Eukaryota) - of currently existing cells.[6]
Using genomics to infer early lines of evolution
Instead of relying a single gene such as the small-subunit ribosomal RNA (SSU rRNA) gene to reconstruct early evolution, or a few genes, scientific effort has shifted to exploiting the comprehensive information from the many complete genome sequences of organisms that are now available.[7]
From such studies, it is now clear that evolututionary trees based only on SSU rRNA alone do not capture the events of early eukaryote evolution accurately, and the progenitors of the first nucleated cells are still uncertain. For instance, careful analysis of the complete genome of the eukaryote yeast shows that many of its genes are more closely related to bacterial genes than they are to archaea, and it is now clear that archaea were not the simple progenitors of the eukaryotes, in stark contradiction to the earlier findings based on SSU rRNA and limited samples of other genes.[8]
One imaginative but disputed hypothesis getting some support from recent computer-assisted studies of complete genome DNA sequences is that the first nucleated cell arose from two distinctly different ancient prokaryotic (non-nucleated) species that had formed a symbiotic relationship with one-another to carry out different aspects of metabolism. One partner of this postulated symbiosis is proposed to be a true bacterial cell, and the other an archaean cell. It is postulated that this symbiotic partnership progressed via the cellular fusion of the partners to generate a chimeric or hybrid cell with a membrane bound internal structure that was the forerunner of the nucleus. The next stage in this scheme was transfer of both partner genomes into the nucleus and their fusion with one-another. Several variations of this hypothesis for the origin of nucleated cells have been suggested.[9]
But some biologists dispute this conception[10] and argue for the need for a shift in conceptual framework if this problem is to be solved. They re-iterate the community metabolism theme, the idea that early living communities would comprise many different entities to extant cells, and would have shared their genetic material more extensively than current microbes.[11]
References
- ↑ Woese C (2002) On the evolution of cells. Proc Natl Acad Sci USA 99:8742-7 PMID 12077305 This article shifts the emphasis in early phylogenic adaptation from vertical to horizontal gene transfer. (Open access.)
- ↑ Koch AL, Silver S.(2005) The first cell. Adv Microb Physiol. 50:227-59. PMID 16221582
- ↑ This theory is expanded upon in The Cell: Evolution of the First Organism by Joseph Panno
- ↑ Kurland CG et al. (2006) Genomics and the irreducible nature of eukaryote cells. Science 312(5776):1011-4 PMID 16709776
- ↑ Hohn, M J 'et al.' (2006). Emergence of the universal genetic code imprinted in an RNA record. Proc. Natl. Acad. Sci. USA 103:18095-18100 PMID 17110438
- O'Donoghue P Luthey-Schulten Z (2003) On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol Mol Biol Rev 67:550–573. PMID 14665676
- Jeffares DC Poole AM Penny D (1998) Relics from the RNA world. J Mol Evol. 46:18-36. Review. PMID 9419222
- Poole AM Jeffares DC Penny D (1998) The path from the RNA world. J Mol Evol. 46:1-17. Review. PMID 9419221
- Gesteland, RF et al. eds.(2006) The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA (2006) (Cold Spring Harbor Lab Press, Cold Spring Harbor, NY,).
- ↑
Olsen G J Woese C R (1997) Archaeal genomics: an overview. Cell 89:991–994 PMID 9215619
- Makarova K S Koonin E V (2005) Evolutionary and functional genomics of the Archaea. Curr. Opin. Microbiol 8:586–594. PMID 16111915
- Woese C R 'et al.' (2000) Aminoacyl-tRNA Synthetases, the Genetic Code, and the Evolutionary Process Microbiol. Mol. Biol. Rev 64:202–236
- Forterre P (2006) Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: A hypothesis for the origin of cellular domain. PNAS 103:3669-3674
- ↑ Daubin V et al. (2003) Phylogenetics and the cohesion of bacterial genomes. Science 301:829-32 PMID 12907801
- Eisen JA, Fraser CM (2003) Viewpoint phylogenomics: intersection of evolution and genomics. Science 300:1706-7 PMID 12805538
- Henz SR et al. (2005) Whole-genome prokaryotic phylogeny. Bioinformatics 21:2329-35 PMID 15166018
- ↑ Esser C et al. (2004) A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol 21:1643-50 PMID 15155797
- Rivera MC, Lake JA (2004) The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431:152-5 PMID 15356622
- Simonson AB et al. (2005) Decoding the genomic tree of life. Proc Natl Acad Sci USA 102 Suppl 1:6608-13 PMID 15851667
- ↑ Esser C et al. (2004) A genome phylogeny for mitochondria among alpha-proteobacteria and a preedominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol 21:1643-50 PMID 15155797
- ↑ Kurland CG et al. (2006) Genomics and the irreducible nature of eukaryote cells. Science 312(5776):1011-4 PMID 16709776
- ↑ Woese C (2002) On the evolution of cells. Proc Natl Acad Sci USA 99:8742-7 PMID 12077305