Human T-lymphotropic virus: Difference between revisions
imported>Sai Kappagantula (New page: {{EZarticle-closed-auto}} ==Classification== Image: Tosco_Refinery.jpg ===Higher order taxa=== Domain; Phylum; Class; Order; family [Others may be used. Use [http://www.tolweb....) |
mNo edit summary |
||
| (57 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{ | {{Subpages}} | ||
{{Taxobox | color=violet | |||
| name = Human T-lymphotropic virus | |||
| image = HTLV-1_1.jpg | |||
| virus_group = Group VI (ssRNA-RT) | |||
| familia = Retroviridae | |||
| genus = Deltaretrovirus | |||
| sero_complex = Human T-lymphotropic virus}} | |||
'''Human T-lymphotropic virus''' was first discovered in Japan in 1977 as the first human [[retrovirus]] to be identified as such. It is thought to be the disease-causing agent in several ailments. [[Paraparesis]] is one disease thought to be caused by the virus where an individual's lower extremities are impaired. The virus is also thought to be an [[oncovirus]], a [[cancer]] causing viral agent. [[Leukemia]], a cancer of [[bone marrow]] or [[blood cells]], has been linked to T-lymphotropic virus. | |||
[[ | |||
==Genome structure== | ==Genome structure== | ||
The virus' genome consists of a single strand of RNA and uses [[reverse transcription]] to form DNA from an RNA template. Among [[retroviruses]] HTLV has a unique genome that leads to its unusual pathogenesis. It shares with other viruses the gag-pol-env motif with flanking LTR (long terminal repeat) sequences. It, however, includes a fourth sequence which acts in an ORF([[open reading frame]]), and leads to products that are most likely pathogenic: Tax, Rex, p12, p13, and p30. | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
HTLV is an [[enveloped virus]]. A core contains the genetic material as well as reverse transcribing proteins that facilitate transcription from RNA to DNA. This core is encased in a protein shell called a [[capsid]]. The outer most portion of the virus is the envelope - a [[phospholipid bilayer]] derived from the host cell that helps the virus invade a host cell. Between the envelope and the capsid is a collection of proteins called the [[tegument]]. | |||
==Ecology== | ==Ecology== | ||
HTLV interacts primarily with human hosts. Close relatives of the virus, however, do exist and invade other animal hosts. | |||
==Pathology== | ==Pathology== | ||
The virus, once in the host, can cause a variety of disease. These diseases include HTLV-1 associated [[myelopathy]], [[opportunistic infections]] from other microbes due to a debilitated immune system, and cancer. In respect to debilitating the immune system, a unique feedback process involving Tax and Rex, an early and rapid replication of the virus is attained. This is followed by a sudden halt to viral replication. The quick replication and abrupt stop sequence allows for the virus to avoid host defenses. This places HTLV in the unique category of [[delta retroviruses]], whose only other members are a few nonhuman viruses: BLV, STLV (simian T-cell leukemia virus), and PTLV (primate T-cell leukemia virus). | |||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
==Current Research== | ==Current Research== | ||
1. Project by The Nyborg Lab of Colorado State University is underway to explore the transciptional regulation of T-cells infected with HTLV. The goal is to study the interactions between the viral proteins, such as the Tax protein, and cellular machinery in vivo and in vitro. ''http://www.nyborglab.com/research.htm'' | |||
2. Study being conducted at Fukuoka University is exploring alternatives to treating Adult T-Cell Leukemia/Lymphoma. Current usage of chemotherapy have insufficient results, but application of stem cell technology has yielded a possible new approach to treating the disease. ''http://www.ncbi.nlm.nih.gov/pubmed/18081707?ordinalpos=6&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum'' | |||
3. Research on the role of nuclear factor-kappaB-inducing kinase using mice with a specific genotype. The study suggests that the lack of NIK inhibits HTLV replication and interrupts the maintenance of the provirus. ''http://www.ncbi.nlm.nih.gov/pubmed/18312467?ordinalpos=3&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum'' | |||
==References== | ==References== | ||
2. http://www.medicalnewstoday.com/articles/95706.php | |||
3. http://www.ncbi.nlm.nih.gov/pubmed/ | |||
4. http://www.emedicine.com/med/topic1038.htm | |||
5. http://www.nyborglab.com/research.htm[[Category:Suggestion Bot Tag]] | |||
Latest revision as of 16:01, 29 August 2024
| Human T-lymphotropic virus | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Virus classification | ||||||||
|
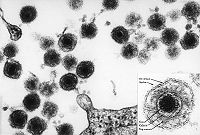
Human T-lymphotropic virus was first discovered in Japan in 1977 as the first human retrovirus to be identified as such. It is thought to be the disease-causing agent in several ailments. Paraparesis is one disease thought to be caused by the virus where an individual's lower extremities are impaired. The virus is also thought to be an oncovirus, a cancer causing viral agent. Leukemia, a cancer of bone marrow or blood cells, has been linked to T-lymphotropic virus.
Genome structure
The virus' genome consists of a single strand of RNA and uses reverse transcription to form DNA from an RNA template. Among retroviruses HTLV has a unique genome that leads to its unusual pathogenesis. It shares with other viruses the gag-pol-env motif with flanking LTR (long terminal repeat) sequences. It, however, includes a fourth sequence which acts in an ORF(open reading frame), and leads to products that are most likely pathogenic: Tax, Rex, p12, p13, and p30.
Cell structure and metabolism
HTLV is an enveloped virus. A core contains the genetic material as well as reverse transcribing proteins that facilitate transcription from RNA to DNA. This core is encased in a protein shell called a capsid. The outer most portion of the virus is the envelope - a phospholipid bilayer derived from the host cell that helps the virus invade a host cell. Between the envelope and the capsid is a collection of proteins called the tegument.
Ecology
HTLV interacts primarily with human hosts. Close relatives of the virus, however, do exist and invade other animal hosts.
Pathology
The virus, once in the host, can cause a variety of disease. These diseases include HTLV-1 associated myelopathy, opportunistic infections from other microbes due to a debilitated immune system, and cancer. In respect to debilitating the immune system, a unique feedback process involving Tax and Rex, an early and rapid replication of the virus is attained. This is followed by a sudden halt to viral replication. The quick replication and abrupt stop sequence allows for the virus to avoid host defenses. This places HTLV in the unique category of delta retroviruses, whose only other members are a few nonhuman viruses: BLV, STLV (simian T-cell leukemia virus), and PTLV (primate T-cell leukemia virus).
Application to Biotechnology
Current Research
1. Project by The Nyborg Lab of Colorado State University is underway to explore the transciptional regulation of T-cells infected with HTLV. The goal is to study the interactions between the viral proteins, such as the Tax protein, and cellular machinery in vivo and in vitro. http://www.nyborglab.com/research.htm
2. Study being conducted at Fukuoka University is exploring alternatives to treating Adult T-Cell Leukemia/Lymphoma. Current usage of chemotherapy have insufficient results, but application of stem cell technology has yielded a possible new approach to treating the disease. http://www.ncbi.nlm.nih.gov/pubmed/18081707?ordinalpos=6&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum
3. Research on the role of nuclear factor-kappaB-inducing kinase using mice with a specific genotype. The study suggests that the lack of NIK inhibits HTLV replication and interrupts the maintenance of the provirus. http://www.ncbi.nlm.nih.gov/pubmed/18312467?ordinalpos=3&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum
References
2. http://www.medicalnewstoday.com/articles/95706.php
3. http://www.ncbi.nlm.nih.gov/pubmed/