Replication of a circular bacterial chromosome: Difference between revisions
imported>Chris Day (→Summary: why the block quote?) |
mNo edit summary |
||
| Line 81: | Line 81: | ||
== References == | == References == | ||
{{reflist|2}} | {{reflist|2}}[[Category:Suggestion Bot Tag]] | ||
Latest revision as of 11:00, 11 October 2024
Unlike the linear DNA of vertebrates, typical bacterial chromosomes contain circular DNA. The duplication of this chromosome prior to cell division is carried out by a complex and efficient set of catalytically active proteins, each dedicated to different tasks needed to replicate a very large molecule in an orderly fashion.
Introduction
A common bacteria that colonizes intestines serves as a excellent example of a bacterium with a circular chromosome. Replication of the Escherichia coli chromosome proceeds in stages, which can be divided into three major headings; initiation, elongation and termination. Bacteria initiate DNA replication at a specific site on the chromosome, the replication origin oriC, from which replication proceeds bidirectionally to the terminus.[1]
Initiation proceeds in a series of well defined biochemical steps, and is the only phase of DNA replication that is known to be regulated, but is regulated such that replication occurs only once in each cell cycle.[2]
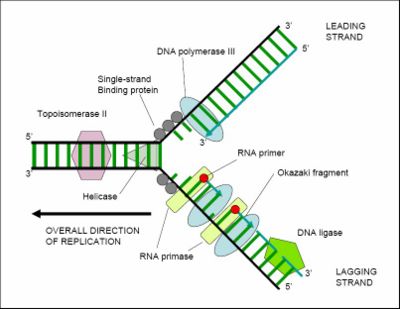
During the elongation phase of replication, two distinct but related events occurs; that is the simultaneous synthesis of the leading and lagging strands. Several enzymes at the replication fork are essential to the synthesis of both strands. Parental strands are first unwound by DNA helicases, and the resulting topological stress is relieved by topoisomerase. Each separated strand is then stabilized by single stranded binding proteins [SSB]. From this point synthesis of leading and lagging strands is very different.
Eventually, the two replication forks of the circular chromosome meet at a terminus region containing multiple copies of a 23 base pair sequence called Ter for terminus.[3] The Ter sequences function as a binding site for a protein called Tus (for Terminus Utilization Substance), whereby replication halts when either replication fork encounters a functional Tus-Ter complex.
Initiation
The E.coli bacterial replication origin, called oriC consists of 245 base pairs bearing DNA sequences that are highly conserved among bacterial replication origins. The chromosomal origin, functions as a site where enzymes assemble to form the machinery that will generate the replication fork.[4]
DNA sequence elements within oriC that are important for its function include DnaA boxes, a 9-mer repeat with a highly conserved consensus sequence 5' - TTATCCACA - 3' [5] , that are recognized by the DnaA protein. DnaA protein plays a crucial role in the initiation of chromosomal DNA replication. [6] Bound to ATP, and with the assistance of bacterial histone-like proteins [HU] DnaA then unwinds an AT-rich region near the left boundary of oriC, which carries three 13-mer motifs [7], and opens up the double-stranded DNA for entrance of other replication proteins.[8]
This region also contains four “GATC” sequences that are recognized by DNA adenine methylase (Dam), an enzyme that modifies the adenine base when this sequence is unmethylated or hemimethylated. The methylation of adenines is important as it alters the conformation of DNA to promote strand separation[9] , and it appears that this region of oriC has a natural tendency to unwind.[10]
DnaA then recruits the replicative helicase, DnaB, from the DnaB-DnaC complex to the unwound region to form the pre-priming complex.[12] After DnaB translocates to the apex of each replication fork, the helicase both unwinds the parental DNA and interacts momentarily with primase .[13]
In order for DNA replication to continue, single stranded binding proteins are needed to prevent the single strands of DNA from forming secondary structures and to prevent them from re-annealing. In addition, DNA gyrase is needed to relieve the topological stress created by the action of DnaB helicase.
Elongation
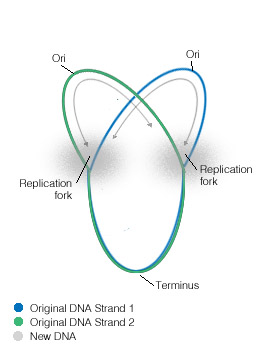
Bacterial chromosomal replication occurs in a bidirectional manner, and has been investigated both genetically and autoradiographically, whereby results provide clear evidence for bidirectional replication in E. coli. In specific experiments, E. coli cells initiating chromosome replication after release from amino-acid starvation were incubated in [3H]thymine of moderate specific activity, followed by incubation in [3H] thymine plus [3H]thymidine of very high specific activity. The grain tracks produced in autoradiographs of chromosomes were denser on both ends than in the middle. The autoradiographic patterns are, therefore, evidence that replication of the chromosome in E. coli is bidirectional.[1]
- See Figure 4 of D. M. Prescott, and P. L. Kuempel (1972): A grain track produced by an E. coli chromosome from cells labeled for 19 min with [3H] thymine, followed by labelingfor 2.5 min with [3H]thymine and ['H]thymidine. [1].[1]
The E. coli replicase DNA polymerase III is a 900 kD complex, possessing a dimeric structure.Each monomeric unit has a catalytic core, a dimerization subunit, and a processivity component .[14] DNA Pol III uses one set of its core subunits to synthesize the leading strand continuously, while the other set of core subunits cycles from one Okazaki fragment to the next on the looped lagging strand. Leading strand synthesis begins with the synthesis of a short RNA primer at the replication origin by the enzyme Primase (DnaG protein).
Deoxynucleotides are then added to this primer by a single DNA polymerase III dimer, in an integrated complex with DnaB helicase. Leading strand synthesis then proceeds continuously, while the DNA is concurrently unwound at the replication fork. In contrast, lagging strand synthesis is accomplished in short Okazaki fragments. First, an RNA primer is synthesized by primase, and, like that in leading strand synthesis, DNA Pol III binds to the RNA primer and adds deoxyribonucleotides.
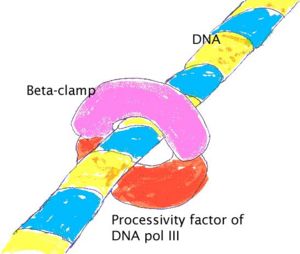
When the synthesis of an Okazaki fragment has been completed, replication halts and the core subunits of DNA Pol III dissociates from the β sliding clamp [B sliding clap is the processivity subunit of DNA Pol III]. [15]The RNA primer is remove and replaced with DNA by DNA polymerase I [which also possesses proofreading exonuclease activity] and the remaining nick is sealed by DNA Ligase, which then ligates these fragments to form the lagging strand.
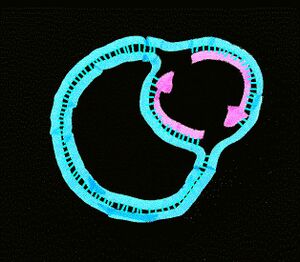
Since replication of a circular chromosome occurs bidirectionally, when the replication fork moves around the circle, a structure shaped like the Greek letter theta Ө is formed. John Cairns provided a well-designed demonstration of E. coli chromosomal replication in 1963. In his experiment, he radioactively labeled the chromosome by growing his cultures in a medium containing 3H-thymidine. The nucleoside base was incorporated uniformly into the bacterial chromosome. He then isolated the chromosomes by lysing the cells gently and placed them on an electron micrograph (EM) grid which he exposed to X-ray film for two months. This Experiment clearly demonstrates the theta replication model of circular bacterial chromosomes.[16]
Termination
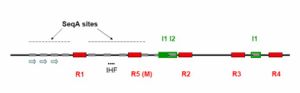
Replication termination of prokaryotic occurs at specific sequences called replication termini.[18] In E.coli, there are 10 replication termini (Ter) located in a region opposite to the replication origin (Fig 5). The Ter sites have polarity, that is, they arrest replication forks, when they are present in one orientation with respect to ori, but allow forks to pass through unrestricted in the opposite orientation. The arrangement of the Ter sites forms a replication trap that forces the two forks, initiated at oriC, to meet each other within a well-defined region of the chromosome.[19]
- See locations and sequences of the replication termini of E. coli.(A) Map showing the ori and the 10 Ter sites. (B) The consensus sequence of Ter. [3][20]
The Ter sites specifically interact with the replication terminator protein called Tus, which is a polar contra-helicase, that is, it impedes the DNA unwinding activity of DnaB in an orientation-dependent manner.[21] The crystal structure of the Tus-Ter complex has been solved, and it reveals a protein that has structural asymmetry and has a DNA-binding domain, consisting of a series of β-strands, that invade the major groove of Ter DNA. The crystal structure was originally interpreted to account for replication fork arrest, solely on the basis of Tus-Ter, protein-DNA interaction, that allegedly was strong enough to form a nonspecific barrier, not only to DnaB helicase-catalyzed DNA unwinding, but in principle, to any protein that would unwind DNA .[22] When either replication fork encounters a functional Tus-Ter complex, it halts replication.
- The crystal structure of the Ter DNA-Tus protein complex (A) showing the nonblocking and the fork-blocking faces of Tus. (B) A cross-sectional view of the helicase-arresting surface.[4][23]
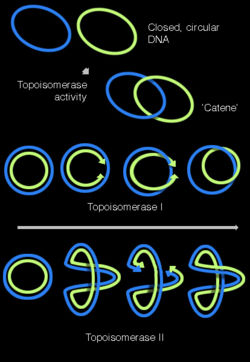
Replication of the DNA separating the opposing replication forks, leaves the completed chromosomes joined as ‘catenanes’ or topologically interlinked circles. The circles are not covalently linked, but cannot be separated because they are interwound and each is covalently closed. The catenated circles require the action of topoisomerases to separate the circles [decatanation]. In E.coli, DNA topoisomerase IV plays the major role in the separation of the catenated chromosomes, transiently breaking both DNA strands of one chromosome and allowing the other chromosome to pass through the break.[24]
There has been some confusion about the role DNA gyrase plays in decatenation. To define the nomenclature, there are two types of topoisomerases: type I produces transient single-strand breaks in DNA and types II produces transient double-strand breaks. As a result, the type I enzyme removes supercoils from DNA one at a time, whereas the type II enzyme removes supercoils two at a time. The topo I of both prokaryotes and eukaryotes are the type I topoisomerase. The eukaryotic topo II, bacterial gyrase, and bacterial topo IV belong to the type II.
We often forget that DNA gyrase does in fact have topoisomerase type II activity; thus, with it being a homologue of topoisomerase IV (also having topoisomerase II activity) we expect similarity in the two proteins' functions. DNA gyrase preliminary role is to introduce negative super coils into DNA, thereby relaxing positive supercoils that come into play during DNA replication. Topoisomerase IV also relaxes positive supercoils, therefore, DNA Gyrase and topoisomerase IV play an almost identical role in removing the positive supercoils ahead of a translocating DNA polymerase, allowing DNA replication to continue unhindered by topological strain. [25]
Confusion arises when some scientific literature state that DNA gyrase is the sole enzyme responsible for decatanation. In an experiment conducted by Zechiedrich,Khodursky and Cozzarelli in 1997, it was found that topoisomerase IV is the only important decatenase of DNA replication intermediates in bacteria.[26] In this particular experiment, when DNA gyrase alone were inhibited, most of the catenanes were unlinked. However, when Topoisomerase IV alone was inhibited, decatenation was almost completely blocked. The results obtained suggest that Topoisomerase IV is the primary decatenase in vivo, and although DNA gyrase does play a role in decatenation, its function is not as essential as topoisomerase IV in the decatentation of interlinked chromosomes.
Conclusion
It is now known that not all bacteria have a single circular chromosome; in fact, some bacteria have multiple circular chromosomes, and many bacteria have linear chromosomes and linear plasmids. However, in bacterial cells which contain a single circular chromosome, such as E.coli, studies have shown DNA replication to occur at specific sequences called the Origin, and proceed bidirectionally in which the partially replicated chromosome forms a theta like structure. Replication continues until the replication fork encounters a functional Tus-Ter complex, which functions as a molecular trap to halt further replication of the molecule. With the assistance of topoisomerase IV in E.coli, the two catenanted circles are then separated forming two double stranded, circular chromosomes.
Summary
Although it is now well established that not all prokaryotic chromosomes exist as a single circular molecule of DNA as previously thought, it still holds true that in most bacterial cells, the chromosome replicates as a circular structure. The synthesis of a DNA molecule can be divided into three stages: initiation, elongation and termination, distinguished by the reactions taking place, and the enzymes involved. In bacterial cells with circular chromosomes, synthesis occurs at the replication fork, the place at which the DNA helix is unwound from the origin until they have copied the whole replicon. When the replication fork moves around the circle, it occurs bidirectionally, and as a result, a structure shaped like the Greek letter theta Ө is formed. Finally since the bacterial chromosome is a single replicon, the fork meets on the other side and two complete double-stranded circular DNA molecules are formed. In this review, we will be focusing on the replication of circular bacterial chromosomes, and we will use a model prokaryote, namely the common gut bacterium Escherichia coli, to demonstrate how circular DNA molecules in prokaryotic organisms are replicated.
Acknowledgments
This is based on an article by Imalda Devaparanam and David Tribe made available under CC by SA licensing conditions from a University course activity at the Department of Microbiology and Immunology, University of Melbourne, 2007.
References
- ↑ 1.0 1.1 1.2 D. M. Prescott, and P. L. Kuempel Bidirectional Replication of the Chromosome in Escherichia coli Proc.Nat.Acad.Sci. USA Vol.69,No.10, pp.2842-2845, October 1972
- ↑ Jon M. Kaguni DnaA: Controlling the Initiation of Bacterial DNA Replication and More.Annu. Rev. Microbiol. 2006. 60:351–71
- ↑ Hill TM, Pelletier AJ, Tecklenburg ML,Kuempel PL. Identification of the DNA sequence from the E. coli terminus region that halts replication forks. Cell. 1988 Nov 4;55(3):459-66.
- ↑ Jon M. Kaguni DnaA: Controlling the Initiation of Bacterial DNA Replication and More.Annu. Rev. Microbiol. 2006. 60:351–71
- ↑ C Weigel, A Schmidt, B Rückert, R Lurz, and W Messer. DnaA protein binding to individual DnaA boxes in the Escherichia coli replication origin, oriC. EMBO J. 1997 November 3; 16(21): 6574–6583.
- ↑ Hirota Y, Mordoh J and Jacob F (1970) On the process of cellular division in Escherichia coli III. Thermosensitive mutants of Escherichia coli altered in the process of DNA initiation. J Mol Biol, 53, 369–387.
- ↑ Bramhill D, Kornberg A. 1988. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 52:743–55
- ↑ Sekimizu K, Bramhill D and Kornberg A (1987) ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E.coli chromosome. Cell, 50, 259–265
- ↑ Gotoh O, Tagashira Y. 1981. Locations of frequently opening regions on natural DNAs and their relation to functional loci. Biopolymers 20:1043–58
- ↑ Kowalski D, Eddy MJ. 1989. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 8:4335–44
- ↑ Jon M. Kaguni DnaA: Controlling the Initiation of Bacterial DNA Replication and More.Annu. Rev. Microbiol. 2006. 60:351–71
- ↑ Carr KM, Kaguni JM. 2001. Stoichiometry of DnaA and DnaB protein in initiation at the Escherichia coli chromosomal origin. J. Biol. Chem. 276:44919–25
- ↑ Tougu K, Marians KJ. 1996. The interaction between helicase and primase sets the replication fork clock. J. Biol. Chem. 271:21398–405
- ↑ O'Donnell M. , Jeruzalmi D. , Kuriyan J. Clamp loader structure predicts the architecture of DNA polymerase III holoenzyme and RFC. Curr. Biol. 11 R935-R946 2001
- ↑ Indiani C, O'Donnell M. Mechanism of the delta wrench in opening the beta sliding clamp. J Biol Chem. 2003 Oct 10;278(41):40272-81. Epub 2003 Jul 8.
- ↑ Cairns, J.P.: Cold Spring Harbor Symposia on Quantitative Biology 28:44, 1963.
- ↑ Cairns, J.P.: Cold Spring Harbor Symposia on Quantitative Biology 28:44, 1963.
- ↑ Bussiere, D. E. & Bastia, D. (1999) Mol. Microbiol. 31, 1611-1618[
- ↑ Sashidhar Mulugu, Aardra Potnis, Shamsuzzaman, Jeffery Taylor, Ken Alexander, and Deepak Bastia. Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. PNAS | August 14, 2001 | vol. 98 | no. 17 | 9569-9574
- ↑ Sashidhar Mulugu, Aardra Potnis, Shamsuzzaman, Jeffery Taylor, Ken Alexander, and Deepak Bastia. Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. PNAS | August 14, 2001 | vol. 98 | no. 17 | 9569-9574
- ↑ Khatri, G. S. , MacAllister, T. , Sista, P. & Bastia, D. (1989) Cell 59, 667-674
- ↑ Kamada, K. , Horiuchi, T. , Ohsumi, K. , Shimamoto, M. & Morikawa, K. (1996) Nature (London) 383, 598-603
- ↑ Sashidhar Mulugu, Aardra Potnis, Shamsuzzaman, Jeffery Taylor, Ken Alexander, and Deepak Bastia. Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. PNAS | August 14, 2001 | vol. 98 | no. 17 | 9569-9574
- ↑ Sashidhar Mulugu, Aardra Potnis, Shamsuzzaman, Jeffery Taylor, Ken Alexander, and Deepak Bastia. Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. PNAS | August 14, 2001 | vol. 98 | no. 17 | 9569-9574
- ↑ Chris Ullsperger and Nicholas R. Cozzarelli .Contrasting Enzymatic Activities of Topoisomerase IV and DNA Gyrase from Escherichia coli. Volume 271, Number 49, Issue of December 6, 1996 pp. 31549-31555
- ↑ E L Zechiedrich , A B Khodursky , N R Cozzarelli. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev. 1997 Oct 1;11 (19):2580-92 9334322