Streptococcus pyogenes: Difference between revisions
imported>Lizbeth Nunez |
imported>Lizbeth Nunez |
||
| Line 34: | Line 34: | ||
==Genome structure== | ==Genome structure== | ||
In 2001 Ferretti et al. published the complete genome of an M1 strain GAS. The genome sequence was determined using whole-genome shotgun approach. The genome is a circular chromosome that consists of 1, 852, 442 bp and contains about 38.5% G+C. One thousand two hundred and eighty two out of 1, 752 ORFs (83%) could be assigned a putative function or have a homolog from another bacterial species. About 615 of these ORfs have an unknown function, 46 are virulence factors, 101 are involved in amino acid transport and metabolism, 109 involved in carbohydrate transport and metabolism, 132 function in translational and ribosomal structure and biogenesis, 79 are stable RNA, 21 function in cell division and chromosome partitioning, 57 in cell envelope biogenesis, outer membrane, 21 in cell motility and secretion, 32 in coenzyme metabolism, 90 in DNA replication, recombination and repair, 58 in energy production and conversion, 172 have a general function prediction only, 48 function in inorganic ion transport and metabolism, 40 in lipid metabolism, 65 in nucleotide transport and metabolism, 38 in posttranslational modification, protein turnover, chaperones, 41 in signal transduction mechanisms, 66 in transcription, 15 are putatively inactive genes. [4] | |||
Group A streptococcus encodes the genes for some stress related proteins, it also has highly conserved genes responsible for osmoregulation. In addition there are genes present that encode proteins associated with “molecular mimicry” (the pathogen can alter its coat so that the host confuses it as its own).[4][6] | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
Revision as of 20:55, 16 April 2008
Articles that lack this notice, including many Eduzendium ones, welcome your collaboration! |
| Streptococcus pyogenes | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [[image:|200px|]] | ||||||||||||||
| Scientific classification | ||||||||||||||
| ||||||||||||||
| Binomial name | ||||||||||||||
| Streptococcus pyogenes |
Description and significance
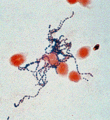
Streptococcus pyogenes is also known as Group A streptococci (GAS). It is a Gram-positive, nonmotile, non-sporeforming coccus Cite error: Closing </ref> missing for <ref> tag.
Growth is optimal on enriched blood agar media but is inhibited if the medium contains a high concentration of glucose [1]. On blood agar plates colonies are usually compact, small and surrounded by a 2-3 mm zone of beta hemolysis (complete lysis of red cells surrounding the colony) that is easily seen and sharply demarcated.
This type of reaction has long been used to classify the streptococci. Group A streptococci is a strict human pathogen, and no other known reservoir or species is affected by diseases unique to this organism [2]
This pathogen is responsible for a vast number of human infections that range from uncomplicated conditions to clinically severe invasive diseases. Some of the diseases are: pharyngitis (most common), tonsillitis (commonly referred to as strep throat), sinusitis, otitis, pneumonia, impetigo, erysipelas, cellulitis, joint or bone infections, necrotizing fasciitis, myositis, meningitis, endocarditis, rheumatic fever and glomerulonephritis (the last two as post streptococcal sequelae) [3][4]. All of these diseases are most common in settings of poverty. By 2005 it was estimated that there were at least 517 000 deaths each year due to severe GAS diseases. They estimated that there are at least 1.78 million new cases each year and a prevalence of at least 18.1 million cases. Most of which are rheumatic heart disease (at least 15.6 million cases). GAS is an important cause of morbidity and mortality [5]
Genome structure
In 2001 Ferretti et al. published the complete genome of an M1 strain GAS. The genome sequence was determined using whole-genome shotgun approach. The genome is a circular chromosome that consists of 1, 852, 442 bp and contains about 38.5% G+C. One thousand two hundred and eighty two out of 1, 752 ORFs (83%) could be assigned a putative function or have a homolog from another bacterial species. About 615 of these ORfs have an unknown function, 46 are virulence factors, 101 are involved in amino acid transport and metabolism, 109 involved in carbohydrate transport and metabolism, 132 function in translational and ribosomal structure and biogenesis, 79 are stable RNA, 21 function in cell division and chromosome partitioning, 57 in cell envelope biogenesis, outer membrane, 21 in cell motility and secretion, 32 in coenzyme metabolism, 90 in DNA replication, recombination and repair, 58 in energy production and conversion, 172 have a general function prediction only, 48 function in inorganic ion transport and metabolism, 40 in lipid metabolism, 65 in nucleotide transport and metabolism, 38 in posttranslational modification, protein turnover, chaperones, 41 in signal transduction mechanisms, 66 in transcription, 15 are putatively inactive genes. [4]
Group A streptococcus encodes the genes for some stress related proteins, it also has highly conserved genes responsible for osmoregulation. In addition there are genes present that encode proteins associated with “molecular mimicry” (the pathogen can alter its coat so that the host confuses it as its own).[4][6]
Cell structure and metabolism
Describe any interesting features and/or cell structures; how it gains energy; what important molecules it produces.
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
Enter summaries of the most recent research here--at least three required
References
- ↑ Murray, Patrick R., Kobayashi, George S., Pfaller Michael A., Rosenthal, Ken S. Medical Microbiology. Second edition. Mosby-Year Book, Inc. St Louis, Missouri 1994.
- ↑ J.J. Ferretti, W.M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, S. Sezate, A. N. Suvorov, C. Primeaux, S. Kenton, H. Lai, S. Lin, Y. Qian, H. Jia, H. Zhu, Q. Ren, F.Z, Najar, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, R. McLaughlin. Complete Genome Sequence of an M1 Strain of Streptococcus pyogenes. PNAS USA. 98, 4658-4663. April 10, 2001.
- ↑ http://www.textbookofbacteriology.net/streptococcus.html
- ↑ Murray, Patrick R., Kobayashi, George S., Pfaller Michael A., Rosenthal, Ken S. Medical Microbiology. Second edition. Mosby-Year Book, Inc. St Louis, Missouri 1994
- ↑ J.R. Carapetis et al., The global burden of group A streptococcal disease, Lancet Infect. Dis. 5 (2005), pp. 685–694.