Bacillus subtilis: Difference between revisions
imported>Annette Kosiorowski |
imported>Annette Kosiorowski |
||
| Line 33: | Line 33: | ||
==Genome structure== | ==Genome structure== | ||
The genome sequence of ''Bacillus subtilis'' was completed and published in November 1997. ''B. subtilis'' contains a single, circular chromosome. It consists of 4,214,810 base pairs. The average G+C content is 43.5%, although ten regions have a G+C content significantly lower than average. There is a significant G-T skew at the third codon positions in common with other bacteria. Its leading strand has a 9% excess of G and 4% excess of T, as compared to the lagging strand. Approximately 87% of the genome consists of protein-coding regions. More than 74% of the all open reading frames and 94% of ribosomal genes are transcribed co-directionally with replication. Only 53% of the genes are represented once. The remaining belong to multigene families, which range in size from 2 to 77 gene copies. Approximately 220 transcriptional regulators have been | The genome sequence of ''Bacillus subtilis'' was completed and published in November 1997. ''B. subtilis'' contains a single, circular chromosome. It consists of 4,214,810 base pairs. The average G+C content is 43.5%, although ten regions have a G+C content significantly lower than average. There is a significant G-T skew at the third codon positions in common with other bacteria. Its leading strand has a 9% excess of G and 4% excess of T, as compared to the lagging strand. Approximately 87% of the genome consists of protein-coding regions. More than 74% of the all open reading frames and 94% of ribosomal genes are transcribed co-directionally with replication. Only 53% of the genes are represented once. The remaining belong to multigene families, which range in size from 2 to 77 gene copies. Approximately 220 transcriptional regulators have been identified in the genome. At the present time, the functions of only approximately 58% of the genes are known, while the functions of 42% are still unknown and being studied. Twelve percent of these genes with unknown functions have homologs in other organisms. | ||
Replication of the ''B. subtilis'' genome proceeds as that of most bacteria, with the splitting of the chromosome into two replication forks. ''B. subtilis'' has an origin of replication (oriC) and a terminus of replication (terC) locate almost perfectly opposite each other on the genome. However, some studies have shown that the clockwise and counterclockwise replication forks actually differ in length by about 170 base pairs. Thus, once the clockwise replication fork reaches the terminus, its synthesis is terminated, while synthesis of the counterclockwise fork continues until it finally reaches the terminus. | Replication of the ''B. subtilis'' genome proceeds as that of most bacteria, with the splitting of the chromosome into two replication forks. ''B. subtilis'' has an origin of replication (oriC) and a terminus of replication (terC) locate almost perfectly opposite each other on the genome. However, some studies have shown that the clockwise and counterclockwise replication forks actually differ in length by about 170 base pairs. Thus, once the clockwise replication fork reaches the terminus, its synthesis is terminated, while synthesis of the counterclockwise fork continues until it finally reaches the terminus. | ||
Revision as of 19:14, 1 April 2008
Articles that lack this notice, including many Eduzendium ones, welcome your collaboration! |
Classification
Higher order taxa
Domain:Eubacteria
Phylum: Firmicutes
Class: Bacilli
Order: Bacillales
Family: Bacillaceae
Species
Genus: Bacillus
Species: subtilis
Description and significance
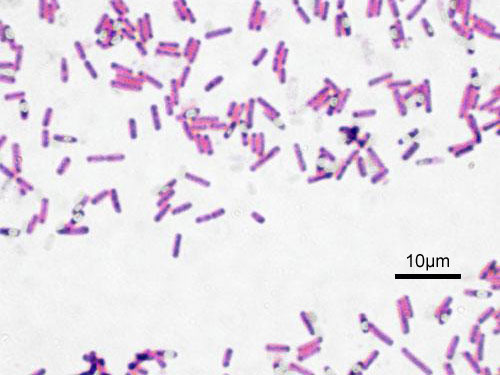
Bacillus subtilis is a gram-positive, rod-shaped, endospore-forming bacteria. It is regarded as an aerobe; however, it is also capable of growing and sporulating under anaerobic conditions when necessary. B. subtilis resides predominantly in soil, including low-nutrient soil. Due to its association with soil particles, it is also inevitably transferred to plants, foods, animals and even marine and freshwater habitats.
B. subtilis was one of the first bacteria studied by scientists. It was originally named Vibrio subtilis in 1835 by Christian Gottfried Ehrenberg and later renamed Bacillus subtilis by Ferdinand Cohn in 1872.
Currently, B. subtilis is a significant microorganism in the scientific research field, as well as in biotechnology and industry. Scientists often utilize B. subtilis as a model organism. B. subtilis becomes naturally competent during its transition between exponential growth and the stationary phase of growth, meaning it is able to bend and internalize DNA from a medium. Thus, it is easily manipulated genetically and a good laboratory microorganism. Due to its endospore-forming mechanisms, it is an especially excellent model system for cell differentiation. Furthermore, B. subtilis produces a variety of enzymes that are used in various industrial roles, including antibiotic production. Its status as a "generally regarded as safe" (GRAS) organism makes it an excellent industrial microorganism.
Genome structure
The genome sequence of Bacillus subtilis was completed and published in November 1997. B. subtilis contains a single, circular chromosome. It consists of 4,214,810 base pairs. The average G+C content is 43.5%, although ten regions have a G+C content significantly lower than average. There is a significant G-T skew at the third codon positions in common with other bacteria. Its leading strand has a 9% excess of G and 4% excess of T, as compared to the lagging strand. Approximately 87% of the genome consists of protein-coding regions. More than 74% of the all open reading frames and 94% of ribosomal genes are transcribed co-directionally with replication. Only 53% of the genes are represented once. The remaining belong to multigene families, which range in size from 2 to 77 gene copies. Approximately 220 transcriptional regulators have been identified in the genome. At the present time, the functions of only approximately 58% of the genes are known, while the functions of 42% are still unknown and being studied. Twelve percent of these genes with unknown functions have homologs in other organisms.
Replication of the B. subtilis genome proceeds as that of most bacteria, with the splitting of the chromosome into two replication forks. B. subtilis has an origin of replication (oriC) and a terminus of replication (terC) locate almost perfectly opposite each other on the genome. However, some studies have shown that the clockwise and counterclockwise replication forks actually differ in length by about 170 base pairs. Thus, once the clockwise replication fork reaches the terminus, its synthesis is terminated, while synthesis of the counterclockwise fork continues until it finally reaches the terminus.
Cell structure and metabolism
Describe any interesting features and/or cell structures; how it gains energy; what important molecules it produces.
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
B. subtilis is not pathogenic to humans, animals, or plants. However, on rare occasions, it may contaminate food and cause food poisoning in humans.
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
Enter summaries of the most recent research here--at least three required
References
Bacillus Subtilis. (2008, March 26). In Wikipedia, The Free Encyclopedia. Retrieved March 26, 2008 from http://en.wikipedia.org/wiki/Bacillus_subtilis