Prokaryote phylogeny and evolution
The fact that genes can move between distant branches of the tree of life even at low probabilities raises challenges to scientists who are trying to reconstruct evolution from studying genes and gene sequences in different organisms, because horizontal gene transfer (HGT) effectively scrambles the information on which biologist are relying to reconstruct a phylogeny of organisms - that is their evolutionary history and relationships. Furthermore, the challenges that are raised by HGT are most awkward for the ambitious (but extremely interesting) reconstruction of the earliest events in evolution - that is the early branches of the tree of life, because over long time intervals and with large numbers of organisms many low probability HGT events are certain to have actually occurred.
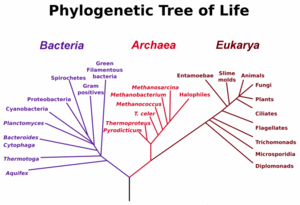
The three main early branches of the tree of life have been intensively studied by microbiologists because the first organisms were microorganisms. Microbiologists (led by Carl Woese) have introduced the term domain for the three main branches of this tree, where domain is a phylogenetic term very similar in meaning to biological kingdom. To reconstruct this tree of life, the sequence of a particular genes encoding the small subunit of ribosomal RNA (SSU rRNA, 16s rRNA) have proven very useful, and the tree shown to the left relies heavily on information from this single gene.
These three domains of life represent the main lineages in evolution of early cellular life and currently represented by the Bacteria, the Archaea (single celled organisms superficially similar to bacteria), and Eukarya (eukaryote) domains.
Eukaryotes are all organisms with a well defined nucleus, and this domain comprises protists, fungi, and all organisms in the animal and plant kingdoms, including humans (See figure at left).
The most common gene used for constructing phylogenetic relationships in microrganisms is the small subunit ribosomal RNA gene, as its sequences tend to be conserved among members with close phylogenetic distances, yet it is variable enough that differences can be measured [1]. The SSU rRNA as a measure of evolutionary distances was pioneered by Carl Woese when formulating the first modern "tree of life", and his results led him to propose the Archaea as a third domain of life.) However, recently it has been argued that SSU rRNA genes can also be horizontally transferred. [2] Although this may be rare, this possibility is forcing scrutiny of the validity of phylogenetic trees based on SSU rRNAs.
Recent discoveries of 'rampant' HGT in microorganisms, and the detection of horizontal movement of even genes for the small subunit of ribosomal RNA have forced biologists to question the accuracy of at least the early branches in the tree shown on the left, and even question the validity of trees as useful models of how early evolution occurs.[3]
"Sequence comparisons suggest recent horizontal transfer of many genes among diverse species including across the boundaries of phylogenetic "domains". Thus determining the phylogenetic history of a species can not be done conclusively by determining evolutionary trees for single genes." [4] HGT is thus a potential confounding factor in inferring phylogenetic trees from the sequence of one gene. For example, if two distantly related bacteria have exchanged a gene, a phylogenetic tree including those species will show them to be closely related even though most other genes have diverged substantially. For this reason it is important to use other information to infer phylogenies, such as the presence or absence of genes, or, more commonly, to include as wide a range of genes for analysis as possible.
Which metaphor: tree, a net, cobweb, or ring?
In his article Uprooting the Tree of Life, W. Ford Doolittle discusses the Last Universal Common Ancestor - the root of the Tree of Life - and the problems with that concept posed by HGT[5]. He describes the microorganism Archaeoglobus fulgidus as an anomaly with respect to a phylogenetic tree based upon the code for the enzyme HMGCoA reductase - the organism is definitely an archaean, with all the cell lipids and transcription machinery expected of an archaean, but its HMGCoA genes are of bacterial origin. In the article, Doolittle says that while it is now widely accepted that mitochondria in eukaryotes derived from alpha-proteobacterial cells and that chloroplasts came from ingested cyanobacteria,
".. it is no longer safe to assume that those were the only lateral gene transfers that occurred after the first eukaryotes arose. Only in later, multicellular eukaryotes do we know of definite restrictions on horizontal gene exchange, such as the advent of separated (and protected) germ cells...
If there had never been any lateral gene transfer, all these individual gene trees would have the same topology (the same branching order), and the ancestral genes at the root of each tree would have all been present in the last universal common ancestor, a single ancient cell. But extensive transfer means that neither is the case: gene trees will differ (although many will have regions of similar topology) and there would never have been a single cell that could be called the last universal common ancestor..."
Doolittle suggested that the universal common ancestor cannot have been one particular organism, but must have been a loose, diverse conglomeration of primitive cells that evolved together. These early cells, each with relatively few genes, differed in many ways, and swapped their genes freely. Eventually, from these eclectic cells came the three domains of life as we know them today: bacteria, archaea and eukaryote. These domains are now recognizably distinct because much of the gene transfer that still occurs is within these domains, rather than between them. Biologist Peter Gogarten reinforced these arguments, and suggested that the metaphor of a tree does not fit the data from recent genome research, and that biologists should instead use "the metaphor of a mosaic to describe the different histories combined in individual genomes and use [the] metaphor of a net to visualize the rich exchange and cooperative effects of HGT among microbes." [6]
Resolution of uncertainty with Phylogenomics

Despite the uncertainties in reconstructing phylogenies back to the beginnings of life, progress is being made in reconstructing the tree of life in the face of uncertainties raised by HGT. The uncertainty of any inferred phylogenetic tree based on a single gene can be resolved by using several common genes or even evidence from whole genomes [8]. One such approach, sometimes called 'multi-locus typing', has been used to deduce phylogenic trees for organisms that exchange genes, such as meningitis bacteria[9].
Jonathan Eisen and Claire Fraser have pointed out that:
"In building the tree of life, analysis of whole genomes has begun to supplement, and in some cases to improve upon, studies previously done with one or a few genes. For example, recent studies of complete bacterial genomes have suggested that the hyperthermophilic species are not deeply branching; if this is true, it casts doubt on the idea that the first forms of life were thermophiles. Analysis of the genome of the eukaryotic parasite Encephalitozoon cuniculi supports suggestions that the group Microsporidia are not deep branching protists but are in fact members of the fungal kingdom. Genome analysis can even help resolve relationships within species, such as by providing new genetic markers for population genetics studies in the bacteria causing anthrax or tuberculosis. In all these studies, it is the additional data provided by a complete genome sequence that allows one to separate the phylogenetic signal from the noise. This is not to say the tree of life is now resolved — we only have sampled a smattering of genomes, and many groups are not yet touched"[10]
These approaches are enabling estimates of the relative frequency of HGT; the relatively low values that have been observed suggests that the 'tree' is still a valid metaphor for evolution - but the tree is adorned with 'cobwebs' of horizontally transferred genes. This is the main conclusion of a 2005 study of more than 40 complete microbial genomic sequences by Fan Ge, Li-San Wang, and Junhyong Kim. They estimate the frequency of HGT events at about 2% of core genes per genome.[11]. Similar whole genome approaches to assessing evolution are also enabling progress in identifying very early events in the tree of life, such as a proposal that eukaryotes arose by fusion of two complete but very diverse prokaryote genomes: one from a bacterium and one from an archaeal cell.[12]
Interestingly, such a fusion of organisms hypothesis for the origin of complex nucleated cells has been put forward by Lynn Margulis using quite different reasoning about symbiosis between a bacterium and an archaen arising in an ancient consortium of microbes. [13]
References
- ↑ Woese C et al (1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". PNAS USA 87: 4576-9. PMID 2112744.
- Woese C, Fox G (1977). "Phylogenetic structure of the prokaryotic domain: the primary kingdoms". PNAS USA 74: 5088-90. PMID 270744.
- ↑ Yap WH et al. (1999) Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonspora chromogena and evidence for horizontal gene transfer of an entire rRNA operon. J. Bacteriol. 181 : 5201-9. PMID 10464188
- ↑ Simonson AB et al. Decoding the genomic tree of life. (2005) Proc Natl Acad Sci U S A. 102 Suppl 1:6608-13. PMID 15851667
- ↑ Horizontal Gene Transfer, Oklahoma State
- ↑ Doolittle WF (2000) Uprooting the tree of life Sci Am 282:90-5 PMID 10710791
- ↑ Gogarten JP 'Horizontal Gene Transfer - A New Paradigm for Biology' PhD thesis
- Zhaxybayeva O, Gogarten JP (2004) Cladogenesis, coalescence and the evolution of the three domains of life Trends in Genetics 20:
- ↑ Ge F et al The cobweb of life revealed by genome-scale estimates of horizontal gene transfer PLoS Biol 3(10), e316
- ↑ Henz SR et al (2005) Whole-genome prokaryotic phylogeny Bioinformatics 21:2329-35 PMID 15166018
- Fitzpatrick DA et al (2006) A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis BMC Evol Biol 6:99
- ↑ Urwin R, Maiden MC (2003) Multi-locus sequence typing: a tool for global epidemiology Trends Microbiol 11:479-87
- Yang Z (2002) Likelihood and Bayes estimation of ancestral population sizes in hominoids using data from multiple loci Genetics 162:1811-23
- Jennings WB, Edwards SV (2005) Speciational history of Australian grass finches (Poephila) inferred from thirty gene trees Evolution Int J Org Evolution 59:2033-47 PMID 16261740
- ↑ Eisen JA, Fraser CM (2003) Viewpoint phylogenomics: intersection of evolution and genomics Science 300:1706-7 DOI: 10.1126/science.1086292
- ↑ Ge F et al (2005) The Cobweb of Life revealed by genome-scale estimates of horizontal gene transfer PLoS Biol 3(10):e316
- ↑ Rivera MC and Lake JA (2004) The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431:152-5 PMID 15356622
- Simonson AB et al. Decoding the genomic tree of life. (2005) Proc Natl Acad Sci U S A. 102 Suppl 1:6608-13. PMID 15851667
- ↑ Acquiring genomes: a theory of the origin of species. Margulis L and Sagan D (2002) Basic Books ISBN 0-465-04392-5